This article requires a subscription to view the full text. If you have a subscription you may use the login form below to view the article. Access to this article can also be purchased.
Graphical Abstract
Abstract
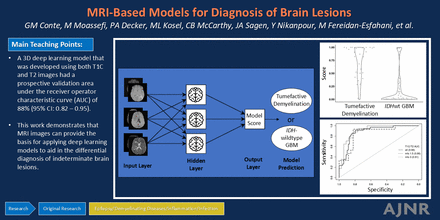
BACKGROUND AND PURPOSE: Diagnosis of tumefactive demyelination can be challenging. The diagnosis of indeterminate brain lesions on MRI often requires tissue confirmation via brain biopsy. Noninvasive methods for accurate diagnosis of tumor and nontumor etiologies allows for tailored therapy, optimal tumor control, and a reduced risk of iatrogenic morbidity and mortality. Tumefactive demyelination has imaging features that mimic isocitrate dehydrogenase wild-type glioblastoma (IDHwt GBM). We hypothesized that deep learning applied to postcontrast T1-weighted (T1C) and T2-weighted (T2) MRI can discriminate tumefactive demyelination from IDHwt GBM.
MATERIALS AND METHODS: Patients with tumefactive demyelination (n = 144) and IDHwt GBM (n = 455) were identified by clinical registries. A 3D DenseNet121 architecture was used to develop models to differentiate tumefactive demyelination and IDHwt GBM by using both T1C and T2 MRI, as well as only T1C and only T2 images. A 3-stage design was used: 1) model development and internal validation via 5-fold cross validation by using a sex-, age-, and MRI technology–matched set of tumefactive demyelination and IDHwt GBM, 2) validation of model specificity on independent IDHwt GBM, and 3) prospective validation on tumefactive demyelination and IDHwt GBM. Stratified area under the receiver operating curves (AUROCs) were used to evaluate model performance stratified by sex, age at diagnosis, MRI scanner strength, and MRI acquisition.
RESULTS: The deep learning model developed by using both T1C and T2 images had a prospective validation AUROC of 88% (95% CI: 0.82–0.95). In the prospective validation stage, a model score threshold of 0.28 resulted in 91% sensitivity of correctly classifying tumefactive demyelination and 80% specificity (correctly classifying IDHwt GBM). Stratified AUROCs demonstrated that model performance may be improved if thresholds were chosen stratified by age and MRI acquisition.
CONCLUSIONS: MRI can provide the basis for applying deep learning models to aid in the differential diagnosis of brain lesions. Further validation is needed to evaluate how well the model generalizes across institutions, patient populations, and technology, and to evaluate optimal thresholds for classification. Next steps also should incorporate additional tumor etiologies such as CNS lymphoma and brain metastases.
ABBREVIATIONS:
- AUROC
- area under the receiver operator curve
- CNSIDD
- CNS inflammatory demyelinating disease
- GBM
- glioblastoma
- IDHwt
- isocitrate dehydrogenase wild-type
- MOGAD
- myelin oligodendrocyte glycoprotein antibody associated disorder
- T1C
- postcontrast T1-weighted
- T2
- T2-weighted
Footnotes
W. Oliver Tobin and Jeanette E. Eckel-Passow are co-senior authors.
This work was supported by the National Institutes of Health (R01NS113803).
Disclosure forms provided by the authors are available with the full text and PDF of this article at www.ajnr.org.
- © 2025 by American Journal of Neuroradiology