Abstract
SUMMARY: In recent years, generative artificial intelligence (AI), particularly large language models (LLMs) and their multimodal counterparts, multimodal large language models, including vision language models, have generated considerable interest in the global AI discourse. LLMs, or pre-trained language models (such as ChatGPT, Med-PaLM, LLaMA), are neural network architectures trained on extensive text data, excelling in language comprehension and generation. Multimodal LLMs, a subset of foundation models, are trained on multimodal data sets, integrating text with another modality, such as images, to learn universal representations akin to human cognition better. This versatility enables them to excel in tasks like chatbots, translation, and creative writing while facilitating knowledge sharing through transfer learning, federated learning, and synthetic data creation. Several of these models can have potentially appealing applications in the medical domain, including, but not limited to, enhancing patient care by processing patient data; summarizing reports and relevant literature; providing diagnostic, treatment, and follow-up recommendations; and ancillary tasks like coding and billing. As radiologists enter this promising but uncharted territory, it is imperative for them to be familiar with the basic terminology and processes of LLMs. Herein, we present an overview of the LLMs and their potential applications and challenges in the imaging domain.
ABBREVIATIONS:
- AI
- artificial intelligence
- BERT
- bidirectional encoder representations from transformers
- CLIP
- contrastive language-image pre-training
- FM
- foundation models
- GPT
- generative pre-trained transformer
- LLM
- large language model
- NLP
- natural language processing
- PLM
- pre-trained language model
- RAG
- retrieval augmented generation
- SAM
- segment anything model
- VLM
- vision language model
The origin of language models dates from the 1990s with statistical language models focused on word prediction using n-grams and hidden Markov models. In 2013, neural language models like Word2Vec shifted the focus to distributed word embeddings using shallow neural networks.1 The field, however, underwent a paradigm shift with the introduction of transformer architecture in 2017, based entirely on attention mechanism.2 This was quickly followed by the introduction of pre-trained language models (PLMs), represented by bidirectional encoder representations from transformers (BERT) (2018) and Bidirectional and Auto-Regressive Transformers (BART) (2019), which marked a major leap by utilizing transformers and context-aware word representations, greatly improving natural language processing (NLP) task performance.3,4 More recently, it was found that scaling PLM (in terms of model or data size), exemplified by generative pre-trained transformer (GPT)-3 (2020) and Pathway Language Models (PaLM) (2022), often leads to not only improved performance on downstream tasks but also some emergent abilities (eg, in-context learning and step-by-step reasoning) in solving a series of complex tasks. To differentiate these language models, the research community introduced the term “large language models” (LLMs) for the PLMs with massive size (eg, containing billions of parameters).5⇓-7
LLMs have rapidly evolved since their introduction. These generative artificial intelligence (AI) models (including LLM, vision language models [VLMs], and diffusion-based models) can generate content in various domains, including language (GPT-4, PaLM, Claude), image (Midjourney, Stable Diffusion), codes (Copilot), and audio (VALL-E, resemble.ai).8⇓-10 Unlike traditional NLP models that process words sequentially, transformer-based models use attention layers to capture long-term dependencies. LLMs are trained on vast data and can produce human-like responses.11 Publicly accessible ChatGPT was initially launched in 2022 by OpenAI12, followed by other LLMs such as Gemini, MedPaLM (Google), LLaVa-Med (Microsoft), Llama (Meta), and Claude 3 (Anthropic). These vary in the training parameters and purposes. While ChatGPT is a general-purpose LLM, MedPaLM and LLaVa-Med, for example, are tailored for medical applications. These models hold promise for enhancing radiology workflows by accelerating information retrieval, generating comprehensive reports, and potentially aiding in diagnostic decision-making.12⇓-14 Given that radiology is often at the leading edge of technology and is associated with more than 70% of FDA-approved AI-enabled tools in the medical domain, it is unlikely that radiologists will remain untouched by this disruptive technology.15
Despite all the excitement around them, current LLMs are still in their infancy. Even though LLMs can mimic human conversations, they rely on word associations rather than true comprehension, limiting their problem-solving and logical reasoning abilities. LLMs may hallucinate or fabricate facts.16 The successful integration of LLMs in radiology demands addressing critical challenges. They need vast training data sets and can give inconsistent responses because of their probabilistic nature. Addressing these challenges is paramount to fully realizing the potential of LLMs in radiology, which can be truly transformative. Herein, we briefly review the evolution of LLMs and their potential applications in radiology, including limitations, challenges, and possible solutions.
Prior to proceeding further, the interested reader is referred to the Supplemental Data for a glossary of common LLM-related terminology.
Models, Modals, and Miscellaneous Things in between!
A foundation model (FM) is an AI model trained using self-supervised learning with large unannotated data sets.17 This confers broad capabilities to the model, enabling it to serve as a base (or foundation) for subsequent models. FMs trained on text, code, and images can be quite versatile. For example, the original ChatGPT (an LLM) was built on GPT 3.5 (a foundation model) and tweaked with chat-specific data.
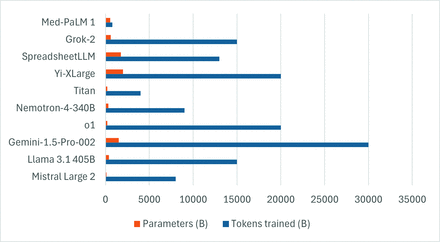
LLMs are a subset of FMs that specialize in language tasks.18 This is done by converting text into “tokens,” which are the fundamental units of input and output data. Tokenization is essential to understanding syntax, semantics, and relationships within the text, affecting eventual model performance and the ability to predict subsequent tokens.19,20 Parameters, on the other hand, are trainable weights and biases within the model, which are learned from the training data and can be considered the building blocks of the model. Figure 1 shows the number of training models and parameters of some of the common LLMs.
Bar chart showing the training tokens and parameters (in billions) of some of the common LLMs. Please note that several models are essentially part of larger families of models, and individual models may have variability in training tokens and parameters (data source: https://lifearchitect.ai/).
Even though LLMs work well for understanding and generating text, these are essentially unimodal, which limits their generalizability. However, integrating image identification/classification is challenging since most deep learning-based computer vision systems are data intensive and broadly not generalizable. While language is discrete and can be tokenized, visual concepts can evolve into higher dimensional spaces and can be difficult to discretize.21 Naively discretizing images on a pixel-by-pixel basis may lose local neighborhood information and may lead to prohibitive computing costs. For example, if the resolution of a color image is 256*256*3, the length of pixel tokens is 196,608. The use of self-attention size (a technique that identifies and weighs the various parts of an input sequence) can scale as a quadratic function of the token length and be computationally prohibitive. This prompted the development of VLMs, which can be broadly defined as multi-modal models capable of performing inference with both images and text. The input may be image or text, while the output can be text, bounding box, or even segmentation masks. As of this writing, there are more than 112 publicly available open-source or application programming interface VLMs, including GPT-4v, Gemini, LLaVA, and others.22 In general, VLMs are trained using four main strategies, either alone or in combination. In contrastive training, pairs of positive and negative examples are used, with the model trained to predict similar representations for the positive pairs. A typical example of this is contrastive learning image pre-training (CLIP), a neural network introduced in 2021.23 CLIP combines a text and an image encoder and leverages information from two modalities to predict which caption goes with which image. The CLIP model has since been used for tasks such as generating images from text (Dalle-3, Midjourney), image segmentation tasks (Segment anything model [SAM]), and tasks involving image captioning and search. Note that CLIP is essentially adept at visual classification tasks. When provided with an image-text pair, it can determine if the two are a good fit for each other. However, it may not work well when differentiating categories with significant overlap.21
The second training strategy for VLMs is masking, where the VLM is trained to reconstruct missing patches in text (given an unmasked image) or vice-versa. The generative training paradigm, on the other hand, is used for models capable of generating entire images or very long captions, although some models may be trained to only generate images from text (eg, stable diffusion).24 These models are generally more expensive to train. Finally, models using pre-trained backbones leverage open-source LLMs to learn the mapping between the image encoder and LLM (eg, Frozen, MiniGPT).21 VLMs may also be further subcategorized into models designed specifically for image interpretation and comprehension in conjunction with language (eg, CLIP), models that generate text from multimodal input (eg, GPT-4V), or models that can have both multimodal input and output (eg, Google Gemini). For a more detailed description of VLM, the interested reader is referred to the recent work by Ghosh et al.8
LLMs may perform the designated task after exposure to a few examples (few-shot learning), single examples (single-shot learning), or even without any training examples (zero-shot learning).5 Another commonly used term in the field is “grounding,” which, in the context of LLMs, essentially implies providing the LLM with relevant and use case-specific information in order to obtain more accurate and relevant output. This is primarily done through retrieval augmented generation (RAG), which retrieves information (through databases, files, etc) and presents it to the LLM, along with the prompt. The LLM then uses this information while responding to the query.25
LLM models may be open-source or proprietary. Open-source LLMs, such as LLaMA series of models, have gained considerable popularity and attention in both academia and industry as a result of the available model checkpoints to customize, transparent model architecture, training process, data sets, and code. In contrast, closed-source LLMs, such as the ChatGPT family, only offer an application programming interface for users to access the LLMs instead of directly using the model. In particular, closed source options may also provide interfaces for users to further fine-tune released models on the host server. In evaluation, although closed-source models often tend to be more powerful because of their access to vast proprietary training data sets and advanced research resources, open-source models are still competitive with top-tier closed-source models.
LLMs, by nature, are probabilistic models, and the response can vary, even to the same query. This, in technical terms, is determined by the “temperature,” which can be adjusted based on the requirements. Models with a higher temperature give a more varied response, which can be entertaining but not ideal in the medical domain. Models with a temperature of zero are deterministic and always give essentially the same response to the same query.26 Some authors have recently proposed using a context-aware temperature network, which can variably drive the temperature up or down, based on the context, eg, TempNet.27 A summary of various LLMs used in the healthcare space is provided in Supplementary Data.
Potential Applications of LLMs in Radiology
LLMs have the potential to impact several facets of radiology, starting from study ordering and protocoling all the way to report generation and follow up. These can impact not only the radiologist but also the patient, primary healthcare providers, and the healthcare system. In the following sections, we briefly outline some of the potential LLM applications and associated challenges.
Workflow Optimization
LLMs, given their ability to comprehend vast textual information (diagnostic requests, electronic health records, prior imaging reports, guidelines, and medical literature)9,28 can help with study protocolling in routine and challenging cases.29 Although tools like ChatGPT and Glass AI have shown promising results in this regard, a study by Nazario-Johnson et al,30 noted that their accuracy currently lags behind that of experienced neuroradiologists.
Chatbots based on LLMs may also be used to provide education about CT/MRI procedures in common terms, thereby reducing patient anxiety while improving patient understanding and engagement.31 Even though LLM responses are generally accurate, they are currently not perfect and require oversight. Also, GPT-4 has been utilized to create summaries and graphical representations of disease courses from the previous MRI reports in patients with glioblastoma, which can potentially save time when comparing multiple prior studies.32
Image Segmentation
FMs can be helpful in reducing the burden associated with manual segmentations, which are labor-intensive and require significant expertise. Unlike deep learning-based semi- or fully automatic segmentation methods, which can have limited generalizability, foundation segmentation models are more broadly generalizable.33 SAM, a segmentation model with zero-shot generalization, generates masks for objects in natural images with distinct boundaries.34,35 Combining SAM with localization algorithms or integrating it with image processing tools like 3D Slicer enhances its medical imaging applications.36 MedSAM is trained on over 1 million medical image-mask pairs from 10 imaging modalities and more than 30 cancer types. MedSAM demonstrates accurate segmentation, achieving results comparable with or better than models like U-Net and DeepLabV3+.37 It could be used for 3D tumor annotation and assessing treatment responses.38 A more recent update, SAM2 is capable of not only segmenting 2Dimages, but also 3D-data sets and videos.39 A more recent addition to the list of segmentation models is CT Foundation, a CT-based model developed for 3D segmentation across different body parts. The model was launched recently by Google and was trained using over a half-million de-identified CT volumes.40 These models could serve as a one-stop shop in the future for radiology-specific tasks instead of having multiple separate segmentation models for individual pathologies (such as glioma, meningioma, and vestibular schwannoma).
Image Interpretation and Report Generation
Some prior studies have noted a superior diagnostic performance of Claude 3 Opus over GPT-4o and Gemini 1.5 Pro in “Diagnosis Please” radiology cases.41 Similarly, GPT-4 Turbo was used to analyze 751 neuroradiology cases from the American Journal of Neuroradiology with an initial diagnostic accuracy of 55.1%, which improved to 72.9% with customized prompt engineering.42 Another recent study compared ChatGPT-4V and Gemini Pro Vision with radiologists and noted 49%, 39%, and 61% accuracy across 190 radiology cases, respectively.43 Finally, models like Bard, ChatGPT-3.5, and GPT-4 have outperformed human consensus and MedAlpaca by at least 5% and 13%, respectively, for rare and complex diagnoses, with GPT-4 achieving a diagnostic accuracy of 93%.44 However, an important caveat here is that all these studies used either history and/or curated limited images per case, as is often the case with online educational content. Unless the radiologist hand-picks individual images for the model to evaluate, along with prompt engineering, the large-scale automated generalization capability of these models is unclear. Similarly, Liu et al45 recently proposed a novel framework for generating radiology reports from high-resolution 3D CT chest data sets without image down-sampling, again showing the potential application of VLMs in 3D-data sets. More simplified LLMs for image interpretation and report generation have also been proposed for 2D images such as chest radiographs.45,46 None of these 2D and 3D models, however, have been extensively evaluated prospectively to ensure fairness, lack of bias, or ability to detect rare diseases, which are important considerations for future validations.
Radiology reports are often written in a freestyle format, which may hinder the extraction of meaningful information for clinical or research purposes.47 LLMs have also been deployed to generate radiology reports by structuring sections such as findings, impressions, and differential diagnoses while integrating demographic data and keywords.48 In a recent study, GPT-4 showed excellent accuracy in selecting the most appropriate report template and identifying critical findings.49 Another study noted AI-structured reports to be comparable to those generated by radiologists, often outperforming the latter in clarity, brevity, and ease of understanding.50
Another retrospective study compared the detection of common reporting errors (eg, omission, insertion, spelling mistakes) by testing a curated data set of erroneous reports. GPT-4 demonstrated a detection rate similar to senior radiologists, attending physicians, and residents (detection rate, 82.7%, 89.3%, 80.0%, and 80.0%, respectively), albeit with reduced time and cost.51 LLMs have also shown promise in automated TNM classification in lung cancer staging based solely on radiology reports without additional training, especially when provided with TNM definitions.52 Finally, LLMs have also shown promise in terms of coding radiology reports and suggesting follow-up based on the presence or absence of pathologies (eg, aortic aneurysm) or employing coding systems (such as Lung-RADS) to screening CT studies.53
Patient-Oriented Reports
LLMs have been shown to simplify radiology report impressions, making them more comprehensible to patients.54 For example, a study comparing LLM-generated MRI spine reports with original reports found that the former had higher comprehension scores among both radiologists and non-physician raters.55 However, these AI-generated reports still require edits and expert supervision, often to remove irrelevant suggestions of causality, prognosis, or treatment.56 Models using patient-friendly language with illustrations of hyperlinked terms perform even better in terms of patient comprehension.55,56 Similarly, some of the recently released LLMs (Med-Flamingo, LLaVA-Med) have shown promising results in visual question answering and rationale generation, which can augment responses to patient queries about reports, implications, and follow-up.57,58
Clinical/Tumor Board Decision-Making in the Future
The role of ChatGPT has also been explored for glioma adjuvant therapy decisions in tumor boards. A small case study (n = 10) noted that the LLM-provided recommendations were rated moderate to good by the experts, even though the model performed poorly in classifying glioma types and lacked sufficient precision to replace expert opinion.59
Radiology Training and Research
LLMs can potentially help simplify complex scientific schematics, compare radiological images, and reduce repetitive tasks and activities that may help in radiology education.60 Another study noted that LLMs like Vicuna-13B can identify various findings on chest radiography reports and show moderate to substantial agreement with existing labelers across data sets like MIMIC-CXR and NIH.26 The LLMs can, therefore, help curate larger data sets while reducing human efforts.
LLMs have also been explored for reviewing manuscripts. The peer review feedback for GPT-4 has shown a considerable overlap (31%) with human reviewers (comparable to overlap between different human reviewers), with users rating it as more beneficial than human feedback.61 LLMs have also been explored for text summarization and editing, especially for non-native authors.62 Use of LLMs for manuscript writing is a big ethical concern and can be a threat not only to the credibility of the paper but also to the authors and the journal itself. Most journals currently do not allow the LLM to be designated as a co-author and ask for transparency from the authors in terms of declaring any use of the LLM in manuscript preparation. Note that LLMs are not databases and are designed to be used as a general reasoning and text engine. They are also well-known to hallucinate references. Some of these problems may be partially overcome with LLMs trained specifically for academic pursuits (eg, Scispace), which can allow the user to interact with pre-selected papers to understand complex research better, extract relevant information, and identify gaps in existing knowledge.63 When used in an ethical way, these resources can potentially enhance the impact of a researcher’s work. Such ethical use is not always a given, and these developments present a more challenging landscape to journals and editors.
LLM Limitations and Drawbacks
Despite the impressive performance of LLMs, there are several limitations and potential risks. A Delphi study highlighted concerns among researchers regarding cybersecurity breaches, misinformation, ethical dilemmas, biased decision-making, and inaccurate communication.19 LLMs generate responses based on statistical pattern recognition, lacking a deep contextual understanding of medical concepts, which can result in errors.64,65 They often fail at common sense reasoning, leading to incorrect or biased outputs. A recent work noted that LLMs can be rather fragile in mathematic reasoning and argued that the current LLMs may not be capable of genuine mathematical reasoning.66 LLMs may produce plausible yet incorrect information “hallucinations” in diagnostics and report generation.67,68
Biases can also arise from failure to capture the complexity of real-world clinical scenarios. This can lead to significant inaccuracies, especially for rare diseases, under-represented groups, third-world populations, and non-English literature. LLMs may perpetuate biases from their training data, leading to misinterpretations and inappropriate treatment recommendations.69 Privacy concerns exist as LLMs are trained on large data sets that may include sensitive patient information. The risk of disclosing such information without consent is considerable.70,71
LLMs can generate convincing but misleading explanations for incorrect answers, known as “adversarial helpfulness.” This can deceive humans and other LLMs by reframing questions, showing overconfidence, and presenting selective evidence. This highlights the need for caution because of the opaque nature of LLMs, which complicates transparency and understanding of their decision-making processes.2,64,72,73 LLMs may struggle to maintain context over long passages, leading to disjointed responses, and they might not be up-to-date with proprietary information or recent advancements.74 LLMs are also vulnerable to adversarial attacks, where malicious inputs deceive the model into producing harmful outputs and reveal confidential information.75
In terms of more specific limitations pertaining to the medical domain, it is unlikely that a single foundational model can serve as a go-to resource given the number of known and continuous newly defined entities, various imaging modalities, and their own inherent resolutions, utilities, and limitations.76 It is also unclear whether a domain-specific LLM versus a modality-specific LLM might be a better long-term solution. Another issue is the lack of high-quality annotations in the medical domain, especially 3D data sets, which limits the amount of available training data. Similarly, the inherent imbalance in the real-world data cannot be ignored. Rare diseases are, by definition, under-represented. Lack of sufficient training data can lead to performance degradation later. Given the dynamic nature of the medical domain, it is inevitable that such models would require continuous retraining and validation. However, occasionally, models may lose previously acquired capabilities while acquiring new ones, as happened with GPT-4 (March 2023), which could differentiate between prime and composite numbers with reasonable accuracy but showed poor performance on the same questions subsequently (GPT-4, June 2023).77 Finally, even though RAG has been shown to alleviate some of the shortcomings of LLMs by grounding and providing context-specific information, the relative prevalence of redundant pieces of information may suppress more recent sparse yet critical information and lead to incorrect responses.78 A hypothetical example would be a recent change in tumor classification or treatment strategy for a certain disease(s). Given the recent change, the information may not be prevalent in the literature and thus be ignored by RAG and LLM while formulating a response. In research, LLMs face limitations due to hallucinations, data bias, misinformation, and a lack of transparency. The training data often lag recent advancements, leading to outdated insights. Also, the resource costs and environmental impact of running LLMs are non-trivial. Over-reliance on these models may erode researchers’ critical thinking and problem-solving skills. Ethical concerns also arise regarding privacy, copyright, and plagiarism, as LLMs cannot be held accountable or listed as authors.79 Consequently, journal guidelines now often require the disclosure of LLM use in manuscript preparation to ensure transparency and maintain the integrity of the review process.80
Future Directions
The US Food and Drug Administration (FDA) has authorized about 1000 AI-enabled medical devices but has yet to authorize an LLM despite acknowledging their potential to positively impact healthcare. Given the complexity of LLMs and the possible output permutations, the FDA recognizes the need for regulatory innovation and specialized tools that allow LLM evaluation in the appropriate context and settings.81 As noted with the various aspects of radiology, the LLM performance currently has considerable challenges in terms of addressing model reliability, explainability, accountability, consistently matching expert-level performance, and withstanding rigorous scrutiny. Even though the news of an LLM outperforming an expert on a test may be eye-catching, radiologists impact several facets of patient care simultaneously in a very dynamic field, and the role of trained medical professionals cannot be taken lightly.
Improving the explainability and generalizability of LLMs is essential for building human trust, given the current limitations.82 Active involvement of domain experts in data selection and model fine-tuning ensures that LLM-generated insights are reviewed and validated before application in patient care, thereby improving accuracy and reducing errors.64 Model training should also address the real-life challenges of imbalanced data and rare diseases, which may considerably impact eventual model performance. This may be done by privacy-compliant data sharing to mitigate real-world data scarcity, limiting the use of synthetic or augmented data (especially for rare cases), and ensuring overall high-quality ground-truth data. It is important to note here that over-reliance on synthetic data can be problematic because it often lacks the complexity of actual data and can lead to model collapse in a real-life setting.83 Patient privacy concerns should be addressed from the training stage itself by excluding any patient-specific identifiers like name, address, and medical record numbers.
The LLM design for the medical domain should also consider the need for tighter regulatory compliances in this field. Models that can provide confidence scores, generating receiver-operating-characteristics curves, are explainable and trained to be fair in terms of patient gender, race, and age and are more likely to survive regulatory scrutiny. Ethical concerns must be addressed to prevent perpetuating biases from training data.69 Compliance with regulations like the Health Insurance Portability and Accountability Act of 1996 (HIPAA) is essential for maintaining data privacy and regulatory compliance.84 Open-source LLMs that can operate locally without sharing data with third parties are a promising privacy-preserving alternative.85 Open-source models (eg, LLaMA) are less reliant on proprietary data sources, potentially increasing transparency and accessibility.14 For proprietary models, additional considerations with regard to the source and quality of training data and any related copyright issues also need to be addressed prior to implementation.
Validating the information generated by LLMs is essential and requires regular audits, fairness-aware training, and ethical guidelines.64,67,86 Paraphrasing a question or providing additional context to an LLM can change the subsequent response.83 Hence, the validation needs to be not only on scientific rigor but also on the contextual understanding of LLM. For example, a rounded peripheral hyper density on a non contrast CT may reflect a contusion (in the context of trauma), a metastatic lesion (in a patient with a known malignant melanoma), a spontaneous hemorrhage (in an older patient with amyloid angiopathy), or a hemorrhagic venous infarct (in a young female on oral contraceptives). Understanding the clinical context in such cases is critical. Traditional scores of accuracy and performance metrics, therefore, may not fully evaluate such models. Similarly, LLM performance may change over time (LLM drift) and can be especially troubling for proprietary models where little is known about the underlying architecture and training data used.83 Additional validation should include testing the LLM on a mix of population cohorts to ensure the model performance is similar with regard to the patient’s gender, demographics, and geographical distribution, ensuring model fairness and lack of bias. LLMs often struggle with outdated information because of static training data. Not only is it important for the LLM to be able to retrieve information from a continuously updated database, but it should also be able to prioritize more recent critical changes over redundant but over-represented literature. Combining LLMs with external data retrieval systems can enhance content generation.87 For example, RadioRAG, a model developed to retrieve real-time information from online resources (like Radiopaedia), variably improved the performance of various LLMs.88 The recent launch of OpenAI o1-preview series aims to tackle complex reasoning tasks by allowing the model to spend more time thinking before responding. This approach helps align it more closely with ethical guidelines and reduces the risk of unsafe or biased content. This model demonstrates expert-level performance on challenging problems by incorporating advanced reasoning capabilities to adhere better to safety protocols and ensure thoughtful, reliable outputs.89
One must also consider the more practical challenges to LLM implementation, including the need for additional energy and infrastructure, resources to ensure continued compliance with regulatory standards, potential safety risks, and medicolegal implications that may offset any efficiency gains. There is also a lack of clarity regarding the cost structure, as some companies may charge based on a number of tokens while others may charge based on hours of usage. Additional costs related to network usage, embedding (use of RAG), and periodic learning will also need to be considered.90 It is also unclear if LLM behavior may change with software or scanner upgrades or the introduction of newer sequences. Finally, establishing mechanisms for fixing liability when an incorrect model decision impacts patient care is also critical and requires both local and national coordination to be uniformly implemented.
In terms of the role of LLMs in neuroradiology, given the uncharted territory, it may be helpful to first validate these models in lower-risk clinical workflows where the LLM output may be annoying or unhelpful but not detrimental to patient health. These may include study protocolling for common exams, summarizing medical history or prior MRI reports, lesion segmentation, and volumetry. Such outputs are open to validation and allow for a more nuanced model evaluation. Using LLMs for lesion characterization or differential diagnosis generation sounds exciting but can pose considerable challenges in real-life settings. It is also important that the model is interpretable. For this, the model would provide not only the possible differential considerations but also the factors that the models considered and how the model weighed them. Another important consideration is rigorous model testing, as model performance may be impacted by model size, domain-specific nature, prompt engineering, and optimization.91 By focusing on these areas in the near future, the field can make early inroads in developing AI solutions that effectively address the complex needs of radiology.
The current challenges in the LLM field also underscore the need for continued short- and long-term research into this field to ensure LLMs are fully utilized. These would include further work into LLMs that are fair, ethical, equitable, and unbiased. Mechanisms that improve model explainability, allow inherent safety guardrails, and minimize or stop hallucinations would further improve user trust. Similarly, further research is needed to find ways for an LLM to continuously update with relevant literature without necessarily forgetting prior information and explore new methods to identify model performance degradation. Equally importantly, further research into new and innovative methods of model validation that use a multi faceted approach beyond traditional performance metrics is needed.
At this time, it is difficult to predict the eventual extent and scale of disruption that LLMs may cause and how they might reshape the role of radiologists in the future. It is possible that LLMs may reduce or eliminate the need for mundane tasks such as study protocolling for common indications, make radiology reports more objective through volumetric inputs, reduce radiologist effort by summarizing impressions, or help with clinical workflows such as patient scheduling, summarizing patient history, and treatment details. LLMs may also play an important role in trainee education, simplifying complex topics, or in research by helping with data collection or annotation. LLMs, in essence, have vast unrealized potential that is dependent on how well the existing challenges are addressed.
CONCLUSIONS
LLMs have transformative potential in radiology with several potential medical applications, but their effective implementation requires addressing key limitations. Researchers and healthcare professionals must navigate these limitations and employ innovative solutions to maximize LLM effectiveness. The future of LLMs in radiology lies in addressing these challenges through interdisciplinary collaboration, ongoing research, and the development of ethical, transparent, and privacy-compliant AI systems. Successful clinical implementation of LLMs would require considerable coordination between domain experts in medicine and computer sciences, researchers, and industry and regulatory authorities.
Footnotes
Disclosure forms provided by the authors are available with the full text and PDF of this article at www.ajnr.org.
Indicates open access to non-subscribers at www.ajnr.org
References
- 1.↵
- 2.↵
- 3.↵
- 4.↵
- 5.↵
- 6.↵
- 7.↵
- 8.↵
- 9.↵
- 10.↵
- 11.↵
- 12.↵
- 13.↵
- 14.↵
- 15.↵
- 16.↵
- 17.↵
- 18.↵
- 19.↵
- 20.↵
- 21.↵
- 22.↵
- 23.↵
- 24.↵
- 25.↵
- 26.↵
- 27.↵
- 28.↵
- 29.↵
- 30.↵
- 31.↵
- 32.↵
- 33.↵
- 34.↵
- 35.↵
- 36.↵
- 37.↵
- 38.↵
- 39.↵
- 40.↵
- 41.↵
- 42.↵
- 43.↵
- 44.↵
- 45.↵
- 46.↵
- 47.↵
- 48.↵
- 49.↵
- 50.↵
- 51.↵
- 52.↵
- 53.↵
- 54.↵
- 55.↵
- 56.↵
- 57.↵
- 58.↵
- 59.↵
- 60.↵
- 61.↵
- 62.↵
- 63.↵
- 64.↵
- 65.↵
- 66.↵
- 67.↵
- 68.↵
- 69.↵
- 70.↵
- 71.↵
- 72.↵
- 73.↵
- 74.↵
- 75.↵
- 76.↵
- 77.↵
- 78.↵
- 79.↵
- 80.↵
- 81.↵
- 82.↵
- 83.↵
- 84.↵
- 85.↵
- 86.↵
- 87.↵
- 88.↵
- 89.↵
- 90.↵
- 91.↵
- Received September 26, 2024.
- Accepted after revision November 13, 2024.
- © 2025 by American Journal of Neuroradiology