Abstract
BACKGROUND AND PURPOSE: Convolutional neural networks are a powerful technology for image recognition. This study evaluates a convolutional neural network optimized for the detection and quantification of intraparenchymal, epidural/subdural, and subarachnoid hemorrhages on noncontrast CT.
MATERIALS AND METHODS: This study was performed in 2 phases. First, a training cohort of all NCCTs acquired at a single institution between January 1, 2017, and July 31, 2017, was used to develop and cross-validate a custom hybrid 3D/2D mask ROI-based convolutional neural network architecture for hemorrhage evaluation. Second, the trained network was applied prospectively to all NCCTs ordered from the emergency department between February 1, 2018, and February 28, 2018, in an automated inference pipeline. Hemorrhage-detection accuracy, area under the curve, sensitivity, specificity, positive predictive value, and negative predictive value were assessed for full and balanced datasets and were further stratified by hemorrhage type and size. Quantification was assessed by the Dice score coefficient and the Pearson correlation.
RESULTS: A 10,159-examination training cohort (512,598 images; 901/8.1% hemorrhages) and an 862-examination test cohort (23,668 images; 82/12% hemorrhages) were used in this study. Accuracy, area under the curve, sensitivity, specificity, positive predictive value, and negative-predictive value for hemorrhage detection were 0.975, 0.983, 0.971, 0.975, 0.793, and 0.997 on training cohort cross-validation and 0.970, 0.981, 0.951, 0.973, 0.829, and 0.993 for the prospective test set. Dice scores for intraparenchymal hemorrhage, epidural/subdural hemorrhage, and SAH were 0.931, 0.863, and 0.772, respectively.
CONCLUSIONS: A customized deep learning tool is accurate in the detection and quantification of hemorrhage on NCCT. Demonstrated high performance on prospective NCCTs ordered from the emergency department suggests the clinical viability of the proposed deep learning tool.
ABBREVIATIONS:
- CNN
- convolutional neural networks
- EDH/SDH
- epidural/subdural hemorrhage
- GPU
- graphics processing unit
- ICH
- intracranial hemorrhage
- IPH
- intraparenchymal hemorrhage
- mask R-CNN
- mask ROI-based CNN
Intracranial hemorrhages (ICHs) represent a critical medical event that results in 40% patient mortality despite aggressive care.1 Early and accurate diagnosis is necessary for the management of acute ICHs.2,3 However, increasing imaging use and distractions from noninterpretive tasks are known to cause delays in diagnosis4 with turn-around time for noncontrast CT head examinations reported up to 1.5–4 hours in the emergency department.4 These delays impact patient care because acute deterioration from hemorrhage expansion often results early, within the initial 3–4.5 hours of symptom onset.5⇓–7 Therefore, a tool for expeditious and accurate diagnosis of ICHs may facilitate a prompt therapeutic response and ultimately improved outcomes.
In addition to ICH detection, a tool for automated quantification of hemorrhage volume may provide a useful metric for patient monitoring and prognostication.8,9 For intraparenchymal hemorrhage (IPH) specifically, the current clinical standard for quantification relies on a simplified formula (ABC/2) calculation that commonly overestimates true IPH volumes by up to 30%.10 Alternatively, while manual delineation of hemorrhage may provide accurate volume estimates, time constraints make this impractical in the emergency setting. Accordingly, a fully automated and objective tool for rapid quantification of ICH volume may be a compelling alternative to current approaches, offering more accurate, detailed information to guide clinical decision-making.
In this study, we propose a tool based on deep learning convolutional neural networks (CNN), an emerging technology now capable of image interpretation tasks that were once thought to require human intelligence.11 The effectiveness of CNNs is based on the capacity of the algorithm for self-organization and pattern recognition without explicit human programming. Using a deep learning approach, Prevedello et al12 previously described a generic algorithm for broad screening of various acute NCCT findings (hemorrhage, mass effect, hydrocephalus) with an overall sensitivity and specificity of 90% and 85%, respectively. We extend this preliminary work by customizing a new mask ROI-based CNN (mask R-CNN) architecture optimized specifically for ICH evaluation and training the network on an expanded cohort of NCCT head examinations. In addition to validation on a retrospective cohort, the trained algorithm will be tested for real-time interpretation of new, prospectively acquired NCCT examinations as part of an automated inference pipeline. By testing performance in a realistic environment of consecutive NCCT examinations, we hope to assess the feasibility of future implementation in clinical practice.
In summary, the 3 key objectives of this study include deep learning algorithm development and assessment of final trained CNN performance in the following: 1) detection of ICH including intraparenchymal, epidural/subdural (EDH/SDH), and subarachnoid hemorrhages; 2) quantification of ICH volume; and 3) prospective, real-time inference on an independent test set as part of an automated pipeline.
Materials and Methods
Patient Selection
After approval of the institutional review board of the University of California, Irvine Medical Center, 2 separate cohorts were identified for this study: one cohort for training (combined with cross-validation) and a second cohort as an independent test set. The initial retrospectively defined training cohort consisted of every NCCT examination acquired at the study institution between January 1, 2017, and July 31, 2017. The subsequent prospectively acquired independent test set cohort consisted of every NCCT examination ordered from the emergency department between February 1, 2018, and February 28, 2018. For both cohorts, cases positive for hemorrhage (IPH, EDH/SDH, and SAH) were identified from clinical reports and confirmed with visual inspection by a board-certified radiologist. 3D ground truth masks were generated for all cases positive for hemorrhage using a custom semiautomated Web-based annotation platform developed at our institution, implementing a variety of tools for level-set segmentation and morphologic operations. All masks were visually inspected for accuracy by a board-certified radiologist.
Convolutional Neural Network
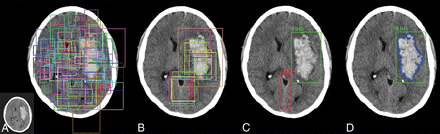
A custom architecture derived from the mask R-CNN algorithm was developed for detection and segmentation of hemorrhage.13 In brief, the mask R-CNN architecture provides a flexible and efficient framework for parallel evaluation of region proposal (attention), object detection (classification), and instance segmentation (Fig 1). In the first step, a preconfigured distribution of bounding boxes at various shapes and resolutions is tested for the presence of a potential abnormality. Next, the highest ranking bounding boxes are identified and used to generate region proposals, thus focusing algorithm attention on specific regions of the image. These composite region proposals are pruned using nonmaximum suppression and are used as input into a classifier to determine the presence or absence of hemorrhage. In the case of detection positive for hemorrhage, a final segmentation branch of the network is used to generate binary masks.
Overview of the mask R-CNN approach. Mask R-CNN architectures provide a flexible and efficient framework for parallel evaluation of region proposal (attention), object detection (classification), and instance segmentation. A, Preconfigured bounding boxes at various shapes and resolutions are tested for the presence of a potential abnormality. B, The highest ranking bounding boxes are identified and used to generate region proposals that focus algorithm attention. C, Composite region proposals are pruned using nonmaximum suppression and are used as input into a classifier to determine the presence or absence of hemorrhage. D, Segmentation masks are generated for cases positive for hemorrhage.
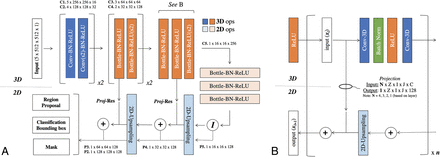
The efficiency of a mask R-CNN architecture arises from a common backbone network that generates a shared set of image features for the various parallel detection, classification, and segmentation tasks (Fig 2). The backbone network used in this article is a custom hybrid 3D/2D variant of the feature pyramid network.14 This custom backbone network was constructed using standard residual bottleneck blocks15 without iterative tuning, given the observation that mask R-CNN architectures, particularly those based on pyramid networks, are robust to many design choices. In this implementation, a 3D input matrix of 5 × 512 × 512 is mapped to 2D output feature maps at various resolutions, with 3D input from the pyramid network bottom-up pathway added to the 2D feature maps of the top-down pathway using a projection operation to match the matrix dimensions. Thus, the network can use contextual information from the 5 slices immediately surrounding the ROI to predict the presence and location of hemorrhage.
Convolutional neural network architecture. A, Hybrid 3D-contracting (bottom-up) and 2D-expanding (top-down) fully convolutional feature-pyramid network architecture used for the mask R-CNN backbone. The architecture incorporates both traditional 3 × 3 filters (blue) as well as bottleneck 1 × 1–3 × 3–1 × 1 modules (orange). The contracting arm is composed of 3D operations and convolutional kernels. Subsampling in the x- and y-directions is implemented via 1 × 2 × 2 strided convolutions (marked by s2). Subsampling in the z-direction is mediated by a 2 × 1 x 1 convolutional kernel with valid padding. The expanding arm is composed entirely of 2D operations. B, Connections between the contracting and expanding arms are facilitated by residual addition operations between corresponding layers. 3D layers in the contracting arm are mapped to 2D layers in the expanding arm by projection operations, which are designed both to match in the input (N) and output (1) z-dimension shape in addition to input (C) and output (128) feature map sizes. Ops indicates operations; Conv, convolutions; BN-ReLU, Batch Normalization Rectified Linear Unit; Proj-Res, Projection-Residual; Z, Z-axis; I, In plane axis; J, In plane axis.
Implementation
The approximate joint training method as described in the original faster mask R-CNN implementation16 was used for parallel optimization of the region-proposal network classifier and segmentation heads. The mask R-CNN architecture was trained using 128 sampled ROIs per image, with a ratio of positive-to-negative samples fixed at 1:3. During inference, the top 256 proposals by the region-proposal network are pruned using nonmaximum suppression and are used to generate detection boxes for classification. The region-proposal network anchors span 4 scales (128 × 128, 64 × 64, 32 × 32, 16 × 16) and 3 aspect ratios (1:1, 1:2, 2:1).
Network weights were initialized using the heuristic described by He et al.17 The final loss function included a term for L2 regularization of the network parameters. Optimization was implemented using the Adam method, an algorithm for first-order gradient-based optimization of stochastic objective functions based on adaptive estimates of lower order moments.18 An initial learning rate of 2 × 10−4 was used and annealed whenever a plateau in training loss was observed.
The software code for this study was written in Python 3.5 using the open-source TensorFlow r1.4 library (Apache 2.0 license; https://github.com/tensorflow/tensorflow/blob/master/LICENSE).19 Experiments were performed on a graphics processing unit (GPU)-optimized workstation with 4 GeForce GTX Titan X cards (12GB, Maxwell architecture; NVIDIA, Santa Clara, California). Inference benchmarks for speed were determined using a single-GPU configuration.
Image Preprocessing
For each volume, the axial soft-tissue reconstruction series was automatically identified by a custom CNN-based algorithm. If necessary, this volume was resized to an in-plane resolution matrix of 512 × 512. Furthermore, all matrix values less than −240 HU or greater than +240 HU were clipped, and the entire volume was rescaled to a range of [−3, 3].
Statistical Analysis
The primary end point of this study was the detection of hemorrhage on a per-study basis. A given NCCT volume was considered positive for hemorrhage if any single region-proposal prediction on any given slice was determined to contain hemorrhage. Thus, algorithm performance including accuracy, sensitivity, specificity, positive predictive value, and negative predictive value was calculated. Furthermore, by varying the softmax score threshold for hemorrhage classification, we calculated an area under the curve.
In addition to complete dataset evaluation, performance statistics on a balanced dataset (an equal number of positive and negative cases) were also calculated. By means of a balanced distribution, accuracy could also be further stratified by hemorrhage type (IPH, EDH/SDH, and SAH) and size (punctate, small, medium, and large, defined as <0.01, 0.01–5.0, 5.0–25, and >25 mL).
The secondary end point of this study was the ability of the algorithm to accurately estimate hemorrhage volume. This was assessed in 2 ways. First, predicted binary masks of hemorrhage were compared with criterion standard manual segmentations using a Dice score coefficient. Second, predicted volumes of hemorrhage were compared with criterion standard annotated volumes using a Pearson correlation coefficient (r). As a comparison, estimates of IPH volume were also calculated using the simplified ABC/2 formula.
Training Cohort Evaluation
A 5-fold cross-validation scheme was used for evaluation of the initial training cohort. In this experimental paradigm, 80% of the data are randomly assigned into the training cohort, while the remaining 20% are used for validation. This process is then repeated 5 times until each study in the entire dataset is used for validation once. Validation results below are reported for the cumulative statistics across the entire dataset.
Independent Test Cohort Evaluation
After fine-tuning the algorithm design and parameters, we applied the final trained network to a new, prospective cohort of all consecutive NCCT examinations ordered from the emergency department for 1 month. The entire pipeline for inference was fully automated, including real-time transfer of newly acquired examinations to a custom GPU server from the PACS, identification of the correct input series, and trained network inference. In addition to initial validation statistics, results from this independent test dataset are also reported.
Results
Patient Selection
The initial training set cohort comprised 10,159 NCCT examinations, 901 (8.9%) of which contained hemorrhage including IPH (n = 358/10,159, 3.5%), EDH/SDH (n = 319, 3.1%), and SAH (n = 224, 2.2%), yielding a total of 512,598 images. The median hemorrhage size was 28.2 mL (interquartile range, 9.4–44.7 mL).
The independent test set cohort compromised 682 prospective NCCT examinations, 82 (12.0%) of which contained hemorrhage including IPH (n = 23, 3.4%), EDH/SDH (n = 38, 5.6%), and SAH (n = 21, 3.1%), yielding 23,668 images. The median hemorrhage size was 24.9 mL (interquartile range, 8.3–35.6 mL). Further baseline stratification of both training and test set cohorts by hemorrhage type and size can be found in Table 1.
Distribution of hemorrhages by type and sizea
ICH Detection
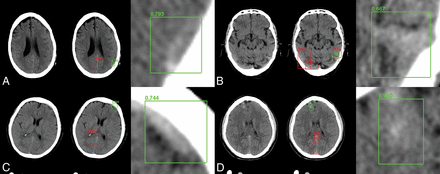
Overall algorithm performance on the full dataset as measured by accuracy, area under the curve, sensitivity, specificity, positive predictive value, and negative predictive value was 0.975, 0.983, 0.971, 0.975, 0.793, and 0.997 for the cross-validation cohort and 0.970, 0.981, 0.951, 0.973, 0.829, and 0.993 for the prospective test set. When stratified by ICH type, the sensitivity for IPH, EDH/SDH, and SAH detection was 98.6% (353/358), 97.4% (311/319), and 94.2% (211/224) for the cross-validation cohort and 100% (23/23), 94.7% (36/38), and 90.5% (19/21) for the prospective test set. In total, 26/901 (2.9%) hemorrhages were missed in the cross-validation cohort compared with 4/81 (4.9%) hemorrhages in the prospective test set (Figs 3 and 4).
Sample network predictions: true-positives. Network predictions by the algorithm include bounding-box region proposals for potential areas of abnormality (to focus algorithm attention) and final network predictions, including confidence of results. Correctly identified areas of hemorrhage (green) include subtle abnormalities representing subarachnoid (A), subdural (B and C), and intraparenchymal (D) hemorrhage. Correctly identified areas of excluded hemorrhage often include common mimics for blood on NCCT, including thickening/high density along the falx (A, C, and D) and beam-hardening along the periphery (B).
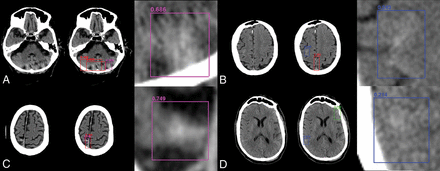
Sample network predictions: false-positives and false-negatives. Network predictions by the algorithm include bounding-box region proposals for potential areas of abnormality (to focus algorithm attention) and final network predictions including confidence of results. False-positive predictions for hemorrhage (purple) often include areas of motion artifacts and/or posterior fossa beam-hardening (A) or high-density mimics such as cortical calcification (C). False-negative predictions for excluded hemorrhage often include small volume abnormalities with relatively lower density, resulting in decreased conspicuity. Examples include subtle subarachnoid hemorrhage along the posterior right frontal lobe (B) and right inferior parietal lobe (D).
Balanced dataset results stratified by hemorrhage size show that in general, algorithm accuracy for hemorrhages of >5 mL (range, 0.977–0.999 mL) is higher than for hemorrhages of <5 mL (range, 0.872–0.965 mL) with only 4 cases of missed hemorrhage of >5 mL across both cohorts (all representing EDH/SDH). Detection accuracy of punctate hemorrhages of <0.01 mL (range, 0.872–0.883 mL) is noticeably more challenging than that of small hemorrhages between 0.01 and 5 mL (range, 0.906–0.965 mL). When we further stratify results by hemorrhage type, the most challenging combinations to detect are punctate SAH or EDH/SDH with accuracy ranges of 0.830–0.881 across both cohorts. Complete stratification of balanced dataset results by hemorrhage and size can be found in Table 2.
Balanced dataset performance statistics stratified by hemorrhage type and sizea
ICH Quantification
Estimates of IPH, EDH/SDH, and SAH segmentation masks by the CNN demonstrated Dice score coefficients of 0.931, 0.863, and 0.772, respectively, compared with manual segmentations. Estimates of IPH, EDH/SDH, and SAH volume by the CNN demonstrated Pearson correlation coefficients of 0.999, 0.987, and 0.953 compared with volumes derived from manual segmentations. By comparison, estimates of IPH volume derived from the simplified ABC/2 formula demonstrated a Pearson correlation of 0.954. On average, the ABC/2-derived hemorrhage volumes overestimated ground truth by an average of 20.2%, while the CNN-derived hemorrhage volumes underestimated ground truth by an average of just 2.1%.
Network Statistics
Each network for a corresponding validation fold trained for approximately 100,000 iterations before convergence. Depending on the number of GPU cards for training distribution, this process required, on average, 6–12 hours per fold. Once trained, the mask R-CNN network was able to determine the presence of hemorrhage in a new test case within an average of 0.121 seconds, including all preprocessing steps on a single GPU workstation.
Discussion
In this study, we demonstrate that a deep learning solution is highly accurate in the detection of ICHs, including IPHs, EDHs/SDHs, and SAHs. In addition, this study demonstrates that a CNN can quantify ICH volume with high accuracy as reflected by Dice score coefficients (0.772–0.931) and Pearson correlations (0.953–0.999). Finally, while embedded for 1 month in an automated inference pipeline, the deep learning tool was able to accurately detect and quantify ICHs from prospective NCCT examinations ordered from the emergency department.
There are several previously described approaches to ICH detection with traditional machine-learning techniques such as fuzzy clustering,20,21 Bayesian classification,22 level-set thresholds,23 and decision tree analysis.24 However, the image diversity present on any given NCCT head examination ultimately limits the accuracy of algorithms that are derived from a priori rules and hard-coded assumptions. For example, Gong et al24 reported a sensitivity of 0.60 and a positive predictive value of 0.447 for IPH detection using decision tree analysis. Furthermore, hard-coded logic tends to produce narrow algorithms optimized for just a single task. For example, Prakash et al23 reported a level-set technique for hemorrhage quantification yielding a Dice score range between 0.858 and 0.917; however, the algorithm is limited for hemorrhage detection because it is not designed to exclude hemorrhage on an examination with negative findings.
Given the increasing awareness of deep learning potential in medical imaging, there has been a gradual paradigm shift increasingly favoring convolutional neural networks over other approaches. For example, Shen et al25 developed a multiscale CNN for lung nodule detection with CT images, while Wang et al26 devised a 12-layer CNN for predicting cardiovascular disease from mammograms as well as for detecting spine metastasis.27 More recently, Phong et al28 described a deep learning approach for hemorrhage detection using several pretrained networks on a small test set of 20 cases.
However, while this preliminary effort is important, there are several key limitations to be addressed before clinical deployment of deep learning tools. First, in addition to high algorithm performance, a clinically viable tool must address the traditional “black box” critique of being unable to rationalize a given interpretation. While there are some techniques to ameliorate this through generation of saliency maps29 or class-activation maps,30 this is a known limitation of conventional global CNN-based classification of an image (or volume). By contrast, the proposed custom mask R-CNN architecture, through combining an attention-based object-detection network with more traditional classification and segmentation components, allows the algorithm to explicitly localize suspicious CT findings and provide visual feedback regarding which findings are likely to represent ICH or a mimic.
Second, a clinically viable tool needs to be tested on unfiltered data in a setting that reflects the expected context for deployment. In this study, we attempted to simulate this by deploying the trained network in a fully automated inference pipeline that can perform all the requisite steps to support algorithm prediction, ranging from PACS image transfer to series identification to GPU-enabled inference, all without human supervision. Furthermore, the prospectively acquired, independent test set used in this context is a reflective sample of the target population used, namely every NCCT head examination performed in the emergency radiology department. That algorithm performance in this setting remains favorable suggests that the deep learning tool has promising potential for clinical utility in the near future.
An additional point should also be made of the requisite data base size for proper algorithm validation. While large datasets are rare in medical imaging, a representative sample of pathology is critical for validating algorithm accuracy. As evidenced in this study, it is often the uncommon findings for which a neural network has the most difficultly learning and generalizing to (eg, punctate hemorrhages of <0.01 mL represent approximately 56/10,841 = 0.5% of all examinations yet are also the most difficult to detect); thus, a large representative dataset is required to assess performance on these critical rare entities. A large data base also facilitates algorithm learning, whereby the increased diversity of training examples helps the network choose more generalizable and predictive features. Finally, cases without ICH are just as important as those with ICH because the algorithm must also be able to correctly identify the absence of hemorrhage in most cases despite any possible underlying pathology that may be present. To address these issues, this study takes advantage of a large training dataset comprising over 512,598 images from >10,000 patients, at least an order of magnitude higher than that in any previous study.
The most salient use case of an accurate tool for hemorrhage detection is a triage system that alerts physicians of examinations potentially positive for hemorrhage for expedited interpretation, thus facilitating reduced turn-around time. The recent 2013 Imaging Performance Partnership survey of >80 institutions rated the importance of reduced turn-around time as one of their highest priorities, scoring 5.7 of a 6.0 rating,31 allowing an expedited triage of patients for therapeutic management. As an example, rapid identification of patients with IPH would facilitate immediate control of blood pressure during the vulnerable first few 3–4.5 hours of symptom onset when acute deterioration is most likely.5⇓–7 The importance of rapid diagnosis is supported further by the recent Intensive Blood Pressure Reduction in Acute Cerebral Hemorrhage Trial-2, which concluded that intensive treatment afforded by early diagnosis was associated with improved functional outcome.32
In addition to hemorrhage detection, ICH volume metrics can be used to precisely and efficiently quantify the initial burden of disease as well as serial changes, which, in turn, may have important clinical implications.33,34 For IPHs, this is most relevant within the first 2–3 hours of onset when the hemorrhagic volume can shift dramatically.5⇓–7 Furthermore, the volume of hemorrhage is a known predictor of 30-day mortality and morbidity.8,9 Presently, the clinical standard for estimation of IPH volume is by the ABC/2 formula of Kwak et al,10,35 in which A and B represent maximum single-dimensional perpendicular measurements on the largest axial region of hemorrhage and C represents a graded estimate of the craniocaudal extent. While easy to use, this limited approach assumes an ellipsoid shape for all IPHs. In this study, we show that this assumption results in overestimation of hemorrhage by 20.2%, a statistic that has been previously reported with discrepancies up to 30% compared with manual segmentation.10 While the criterion standard remains manual delineation, this approach can be both time-consuming and technically challenging in the emergency department setting. By comparison, the ability of the trained CNN to rapidly and accurately quantify IPH volume with >0.999 correlations to human experts offers a clinically feasible, improved alternative to the current standards of practice.
Several limitations should be addressed when considering our results. First, examinations in this study were performed at a single academic institution. Therefore, while we have demonstrated that our results generalize well to independent datasets obtained at our hospital center, further work is necessary to evaluate performance on a variety of vendors and scanning protocols at other institutions. While we acknowledge this drawback, CT examinations are inherently normalized by Hounsfield Units and show less image variability than plain radiographs or MR imaging. Second, deep learning algorithms are known to be susceptible to the phenomenon of adversarial noise,36 where small but highly patterned perturbations in images may result in unexpected predictions. However, this is rare and was not encountered in the current dataset and, to some extent, can be mitigated using network ensembles and denoising autoencoders.37 Finally, while the current dataset is quite large, there are, nonetheless, rare findings and contexts that occur at a prevalence of less than our 1/10,000 cases, and it is foreseeable that such studies may be incorrectly interpreted. To this end, we plan to incorporate continued iterative algorithm updates as new, increasingly larger datasets become available.
Conclusions
This study demonstrates the high performance of a fully automated, deep learning algorithm for detection and quantification of IPH, EDH/SDH, and SAH on NCCT examinations of the head. Furthermore, confirmation of high algorithm performance on a prospectively acquired, independent test set while embedded in an automated inference environment suggests the clinical viability of this deep learning tool in the near future. Such a tool may be implemented either as a triage system to assist radiologists in identifying high-priority examinations for interpretation and/or as a method for rapid quantification of ICH volume, overall expediting the triage of patient care and offering more accurate, detailed information to guide clinical decision-making.
Footnotes
Disclosures: Peter Chang—RELATED: Grant: National Institutes of Health (National Institute of Biomedical Imaging and Bioengineering) T32 Training Grant T32EB001631.* Christopher G. Filippi—UNRELATED: Consultancy: Syntactx, Comments: I read MR imaging brain scans for their clinical trials; Grants/Grants Pending: The Foundation of the American Society of Neuroradiology (FASNR), Comments: Alzheimer research study grant*; Payment for Lectures Including Service on Speakers Bureaus: Grand Rounds Lectures: Montefiore, Albert Einstein College of Medicine, and Iowa University. Daniel Chow—RELATED: Grant: Canon Medical. *Money paid to the institution.
The work of P.C. was, in part, supported by grant T32EB001631 from the National Institutes of Health (National Institute of Biomedical Imaging and Bioengineering). The work of D.C. was, in part, supported by Canon Medical Systems.
Indicates open access to non-subscribers at www.ajnr.org
References
- Received April 10, 2018.
- Accepted after revision June 6, 2018.
- © 2018 by American Journal of Neuroradiology