Abstract
BACKGROUND AND PURPOSE: Accurate segmentation of brain metastases is important for treatment planning and evaluating response. The aim of this study was to assess the performance of a semiautomated algorithm for brain metastases segmentation using Background Layer Statistics (BLAST).
MATERIALS AND METHODS: Nineteen patients with 48 parenchymal and dural brain metastases were included. Segmentation was performed by 4 neuroradiologists and 1 radiation oncologist. K-means clustering was used to identify normal gray and white matter (background layer) in a 2D parameter space of signal intensities from postcontrast T2 FLAIR and T1 MPRAGE sequences. The background layer was subtracted and operator-defined thresholds were applied in parameter space to segment brain metastases. The remaining voxels were back-projected to visualize segmentations in image space and evaluated by the operators. Segmentation performance was measured by calculating the Dice-Sørensen coefficient and Hausdorff distance using ground truth segmentations made by the investigators. Contours derived from the segmentations were evaluated for clinical acceptance using a 5-point Likert scale.
RESULTS: The median Dice-Sørensen coefficient was 0.82 for all brain metastases and 0.9 for brain metastases of ≥10 mm. The median Hausdorff distance was 1.4 mm. Excellent interreader agreement for brain metastases volumes was found with an intraclass correlation coefficient = 0.9978. The median segmentation time was 2.8 minutes/metastasis. Forty-five contours (94%) had a Likert score of 4 or 5, indicating that the contours were acceptable for treatment, requiring no changes or minor edits.
CONCLUSIONS: We show accurate and reproducible segmentation of brain metastases using BLAST and demonstrate its potential as a tool for radiation planning and evaluating treatment response.
ABBREVIATIONS:
- BL
- background layer
- BLAST
- Background Layer Statistics
- BM
- brain metastases
- DL
- deep learning
- DSC
- Dice-Sørensen coefficient
- HD
- Hausdorff distance
- ICC
- intraclass correlation coefficient
- IQR
- interquartile range
- SRS
- stereotactic radiosurgery
- TH
- threshold
Brain metastases (BM) are diagnosed in up to 40% of patients with metastatic cancer and usually imply a short survival.1 However, recent advances in the treatment of BM, with, for example, stereotactic radiosurgery (SRS) have led to improvements in patient outcomes with less impact on neurocognition and quality of life.2 A requirement for SRS is accurate detection and contouring of BM. Additionally, treatment of BM requires measurements of tumor burden at baseline and follow-up to assess treatment response. To aid in this requirement, accurate segmentation of the tumor is needed to provide precise lesion targeting and to monitor changes in the size of the metastases between baseline and follow-up scans.
During the past few years, advances in machine learning methods have led to improvements in automated and semiautomated brain tumor segmentation. Machine learning methods can be grouped into supervised and unsupervised algorithms. Supervised methods, such as those based on deep convolutional neural networks, have recently garnered attention, showing excellent performance in brain tumor segmentation tasks,3⇓-5 with 1 algorithm now FDA-cleared.6 These supervised methods, however, require large numbers of (manual) labels for training, which is a time-consuming and costly process and can be prone to bias introduced by the training set. Additionally, optimal performance of deep learning (DL) algorithms across multiple institutions commonly requires retraining with additional site-specific data (distributions).7
Unsupervised techniques do not require a priori training and can be used to facilitate the creation of ground truth data, which can be used to train DL models. Unsupervised techniques using clustering methods (eg, K-means, fuzzy c-means, and the expectation-maximization method) are iterative algorithms that segment by grouping voxels with similar signal properties (intensities), then estimating and optimizing cluster properties.8 K-means clustering groups signal intensity data into k classes by iteratively computing a mean intensity for each class and clustering voxels into the closest class centroid. Brain tumor segmentation with these techniques can be challenging because segmentation performance is highly dependent on initial conditions and the algorithm used.9 Furthermore, the signal heterogeneity and the small size of BM relative to background brain are additional challenges for clustering algorithms, to accurately identify and classify these tumors.
In this article, we describe an alternative semiautomated method for segmentation of BM using multiparametric MR imaging. The methodology first establishes a parameter space with origin and axes defined by the signal intensity statistics of background brain (Background Layer Statistics [BLAST]). In the present implementation of the methodology, K-means clustering is used to define the statistics of the background layer on a section of normal brain. Within the parameter space, voxels related to the background layer are then removed from the entire volume and additional operator-defined thresholds are applied to preferentially detect and segment BM. In this study, we evaluate BLAST methodology for segmentation of BM and hypothesize that it is accurate and reproducible.
MATERIALS AND METHODS
Subjects
Consecutive patients with newly diagnosed BM undergoing pretreatment MR imaging from July 1, 2022, to September 30, 2022, were included in this retrospective study. Research ethics board approval was obtained at our institution (Sunnybrook Health Sciences Centre, Toronto, Ontario, Canada). Inclusion criteria were the following: 1) known biopsy-proved primary malignancy at the time of brain MR imaging, 2) 10 or fewer BM, and 3) no relevant treatment history at the time of brain MR imaging, including chemotherapy, radiation therapy, or prior brain surgery. Exclusion criteria were the following: 1) the presence of leptomeningeal disease or hemorrhagic metastasis, 2) the presence of another coexisting acute process such as acute stroke, or 3) severe corruption of MR imaging by motion artifacts.
MR Imaging Acquisition
All imaging was performed on 1.5T (Magnetom Aera or Sola; Siemens) or 3T (Magnetom Vida; Siemens) MR imaging systems using body-transmit and 20-channel head and neck receiver coils. Patients were scanned with the institutional brain tumor imaging protocol including axial RESOLVE DWI (Siemens) (b-values = 0 and 1000 s/mm2, TR = 3650–8010 ms, TE = 67.2–72.2 ms, in-plane resolution = 0.54 × 0.54 mm2 to 1.25 × 1.25 mm2, section thickness = 5 mm), axial T2 FLAIR postgadolinium (TR = 9000 ms, TE = 80–108 ms, TI = 2500 ms, in-plane resolution = 0.75 × 0.75 mm2 to 0.83 × 0.83 mm2, section thickness = 3 or 5 mm), and 3D T1 MPRAGE postgadolinium (TR = 1800 or 2240 ms, TE = 2.4 or 3 ms, flip angle = 8°, resolution = 1 × 1 × 1 mm3).
Image Processing and Analysis
T2 FLAIR and trace DWI (b=1000 s/mm2) images were registered to 3D T1 MPRAGE using BRAINSFit (3D Slicer; https://www.slicer.org/)10 with a 6-df rigid registration and linear interpolation to 1-mm isotropic resolution. Brain extraction was performed using a custom-written script in Matlab and the Image Processing Toolbox, Release 2022a and 2023a (MathWorks) which uses a brain mask derived from a binary threshold of the coregistered and interpolated trace DWI (b=1000 s/mm2) data set. Brain-extracted T2 FLAIR and 3D T1 MPRAGE data were bias-field corrected using the N4ITK algorithm (3D Slicer)10 and saved in a NIfTI format.
Ground truth segmentations of BM were manually acquired on the 3D T1 MPRAGE images (ITK-SNAP; www.itksnap.org)11 by C.H., a neuroradiologist with 8 years of postfellowship experience. All ground truth segmentations were reviewed and edited by H.S., a neuroradiation oncologist with 13 years of postfellowship experience. The ROI was drawn around each metastasis encompassing the entire enhancing portion of the brain metastasis as well as central areas of necrosis and excluding edema, generating a binary mask (for a 1-class segmentation task). For this study, enhancing was defined as having a qualitative signal intensity above the surrounding brain parenchyma on the postgadolinium T1 MPRAGE sequence.
BLAST
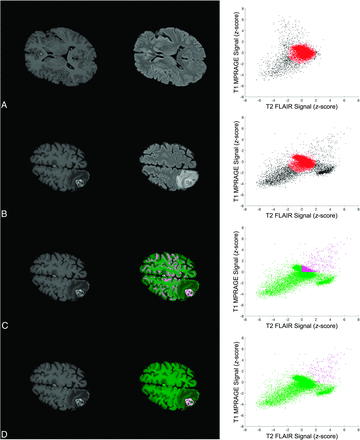
Figure 1 illustrates the key concepts in defining the BLAST parameter space and performing segmentations of enhancing tumor. An example of segmentation of enhancing tumor with vasogenic edema is shown in the Online Supplemental Data. The algorithm was implemented using a custom-written script in Matlab. Signal intensities of the skull-stripped, coregistered, and bias-corrected T1 MPRAGE and T2 FLAIR images were first rescaled from 0 to 1. On a section of normal brain, K-means clustering (2 clusters) of T1 MPRAGE and T2 FLAIR signal intensities was used to define the cluster corresponding to normal gray and white matter (background layer). For this study, the normal section (without any evidence of enhancing tumor or edema) was selected beforehand by the investigators. The centroid of this background layer cluster was then used to define the origin of the parameter space. The rescaled signal intensities in T1 MPRAGE and T2 FLAIR were divided by the SD of the background layer to redefine the position of data points in this parameter space as a z score.
Key steps in defining the BLAST parameter space and performing segmentations with BLAST. Images and segmentations are shown in the left and middle columns, and the corresponding parameter space is shown in the right column. Each point in the scatterplot represents a voxel in the image. A, The statistics of the background layer (gray and white matter) are first defined using K-means clustering on a normal section of the brain. The origin of the parameter space is defined by the centroid of the background layer cluster and the axes expressed as z score relative to the background layer cluster SD. B, An ellipsoid approximating the background layer is applied to all slices (red ellipse). Voxels corresponding to enhancing brain tumor, edema, and CSF are found in the upper right, lower right, and lower left corner of parameter space, respectively. C, Thresholds in both T1 MPRAGE and T2 FLAIR are applied to exclude nontumoral voxels to perform segmentations. In the sample shown, thresholds in both parameters are set to the mean of the background layer (z score = 0). D, Voxels falling within the background layer ellipsoid are subtracted to further exclude the background layer. The segmentation is selected to save a mask of the 3D-connected enhancing BM voxels. An algorithm to fill in necrotic areas is applied for the final segmentation (not shown).
Once signal intensities were put into the BLAST parameter space, segmentations were performed by subtracting the background layer and applying additional thresholds in T1 MPRAGE and T2 FLAIR to exclude other nontumor tissues such as blood vessels and dura. Background layer subtraction was achieved by approximating the background layer voxels using an ellipsoid with the center set to the origin of the BLAST parameter space. The remaining voxels in parameter space were projected back into the image space to visualize the resulting segmentation of enhancing tumor. The 3D object consisting of connected enhancing voxels was selected, and an algorithm to “fill in” nonenhancing or necrotic regions was applied to finalize the segmentation. This filling was completed using morphologic dilations and erosion with structuring elements. To produce an outline of the brain metastasis, we applied a line contour algorithm to encompass the segmentation.
Ablation Study
An ablation study was performed to evaluate the contribution of different steps to segmentation performance. The complete BLAST method consisted of application of thresholds in 2D (T1 MPRAGE and T2 FLAIR) and subtraction of the background layer approximated by an ellipsoid with the semiaxis length set to 1.5 (threshold [TH] × 2 BL = 1.5). Algorithms consisting of application of thresholds in 2D without background layer subtraction (TH × 2 background layer [BL] = 0) and an algorithm consisting of a threshold in 1D (T1 MPRAGE) without background layer subtraction (TH × 1 BL = 0) were also evaluated. For TH × 2 BL = 1.5 and TH × 2 BL = 0, the thresholds in T1 MPRAGE were incremented by 0.5 from the z score = −1 up to z score = 3, and those in T2 FLAIR were incremented by 0.5 from the z score = −2 up to z score = 3. For TH × 1 BL = 0, the threshold in T1 MPRAGE was iterated from the z score = −1 up to z score = 5 by increments of 0.5. For each iteration, the segmentation result was saved and the Dice-Sørensen coefficient (DSC) for the tumor volumes was calculated in Matlab using the manual segmentations as the ground truth.
Evaluation of Background Layer Subtraction on Segmentation Performance
To understand the effect of background layer subtraction on segmentation performance, we ran the algorithm with different amounts of background subtraction by varying the ellipsoid semiaxis length from 0 to 3 by increments of 0.5. This step was performed for thresholds in T1 MPRAGE and T2 FLAIR of 0, 0.5, and 1. For each combination, the segmentation result was evaluated by calculating the DSC.
Operator-Generated BLAST Segmentations and Contours
BM segmentation was performed by 5 operators (4 board-certified neuroradiologists with BLAST using background layer subtraction and thresholding in 2D [TH × 2 BL = 1.5]: E.W., T.K., A.K., and, P.H., with 1–15 years of postfellowship experience and 1 board-certified neuroradiation oncologist: C.-L.T with 8 years of postfellowship experience treating and contouring BM). All definitive BM were identified by the principal investigators, and the location was provided to the operators. Operators segmented all BM within a patient including dural-based metastases. Bone metastases were excluded because these were removed from the volume by the brain-extraction algorithm.
The size of the metastases was estimated as the length of the major axis of an ellipsoid with the same normalized second central moment as the segmented 3D object. Volume was calculated from the number of voxels comprising the segmented 3D object. In addition to the DSC, the Hausdorff distance (HD) for the center section of a metastasis was calculated using Matlab.
To evaluate the clinical acceptance of the BLAST-generated contours, we used a 5-point Likert scale adapted from the MD Anderson Cancer Center.12 Scoring was performed by an experienced neuroradiation oncologist (H.S.) using the contour from the operator with the highest DSC for the case. A score of 4 or 5 on the Likert scale, for example, indicates that the contour is acceptable for clinical practice, while a score of 2 indicates that the contour is completely unusable.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism software, Version 9.5.1 for Mac OS (GraphPad Software). The Shapiro-Wilk test was used to test for normality. For nonparametric group comparison, the Mann-Whitney U test or Friedman test was performed, and the results were considered significant if the P value was less than .05. A linear regression of tumor volume measured by BLAST segmentation versus ground truth segmentation was performed. The difference in tumor volume between BLAST and ground truth segmentation was also measured with a Bland-Altman plot.
Interreader agreement of tumor volume measurements was evaluated by a 2-way random effects model intraclass correlation coefficients (ICC) in Matlab:13 ICC < 0.5 (poor), 0.5–0.75 (fair), 0.75–0.9 (moderate), and ≥0.9 (excellent) agreement. ICCs were reported with their 95% CIs.
RESULTS
A total of 19 patients with 48 BM met the inclusion and exclusion criteria (Table). The median volume of BM was 0.70 cm3 (interquartile range [IQR], 0.1–2.1 cm3) and the median diameter was 12.8 mm (IQR, 6.7–20.7) mm. Twenty-two BM measured <10 mm, and 6 BM measured <5 mm.
Patient demographics and tumor characteristics
Results from the ablation experiment are included in the Online Supplemental Data. The median DSC scores for TH × 2 BL = 1.5, TH × 2 BL = 0, and TH × 1 BL = 0 were 0.9 (IQR, 0.87–0.92), 0.90 (IQR, 0.88–0.92), and 0.86 (IQR, 0.78–0.9), respectively. DSC scores for TH × 2 BL = 0 and TH × 2 BL = 1.5 were significantly higher than those for TH × 1 BL = 0 (P < .001). DSC was not significantly different between TH × 2 BL = 0 and TH × 2 BL = 1.5 (P = .94). An illustrative case in which TH × 1 BL = 0 failed to properly segment a brain metastasis abutting the adjacent tentorium is also shown.
Results of varying the levels of background layer subtraction on segmentation performance are included in the Online Supplemental Data. The experiment was run on 46 of the 48 BM. Two BM demonstrated low T2 FLAIR signal, and the signal intensities for the tumor fell below the minimum T2 FLAIR (z score = 0) used in the experiment. For a given threshold in T1 MPRAGE and T2 FLAIR, increasing background layer subtraction by varying the semiaxis length from 0 to 3 resulted in improving segmentation performance. The benefit of background layer subtraction is more pronounced for lower thresholds and reduces at higher thresholds in T1 MPRAGE and T2 FLAIR. Visually, background layer subtraction reduces the amount of background voxels included in the segmentation mask, allowing better visualization of metastases across the thresholds that were evaluated. While there is greater background layer removal at higher thresholds or with greater background layer subtraction, there is also reduction in the tumor segmentation.
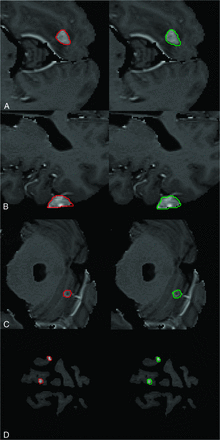
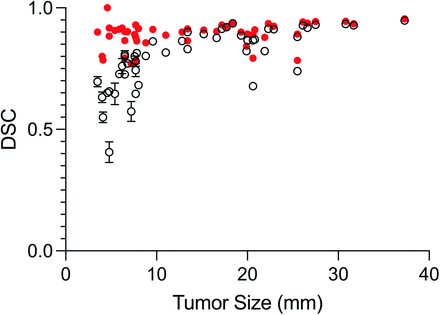
Sample contours for a cross-section of metastases contoured by human operators are shown in Fig 2. Figure 3 shows the relationship between DSC versus metastasis size for the operators and compares this with the results from the ablation analysis (TH × 2 BL = 1.5). The median DSC for all BM and all operators was 0.82 (IQR, 0.73–0.9). For operators, there was a significant difference in DSC (P < .001) between metastases of <10 mm (0.70; IQR, 0.65–0.8) and those of ≥10 mm (0.90; IQR, 0.86–0.92). A statistically significant difference in the median DSC for metastases of <10 mm (0.90; IQR, 0.84–0.91) and those of ≥10 mm (0.91; IQR, 0.89–0.94) was not found for TH × 2 BL = 1.5 from the ablation experiment (P = .05). The median HD for all metastases and operators was 1.4 mm (IQR, 1–2 mm). The median HD was significantly different (P < .001), for metastases of <10 mm (1.4 mm; IQR, 1–1.4 mm) and those of ≥10 mm (1.8 mm; IQR, 1.1–2.2 mm).
Sample contours derived from BLAST segmentations for parenchymal and dural metastases. The first column shows ground truth segmentations for 5 metastases (red). Corresponding contours created with BLAST are shown in the second column (green) for the operator with the best DSC. The mean DSCs for A, B, and C are 0.90, 0.83, 0.80, respectively. Two small metastases measuring 7 and 8 mm are shown in D with DSCs of 0.67 and 0.82, respectively.
BLAST segmentation performance measured by DSC (mean) as a function of metastasis size for all operators (black open circles). Segmentation performance for BLAST generated by automatically iterating through combinations of thresholds with fixed background layer subtraction (red closed circles) outperforms operator-generated segmentations for small (<10 mm) metastases (P < .001).
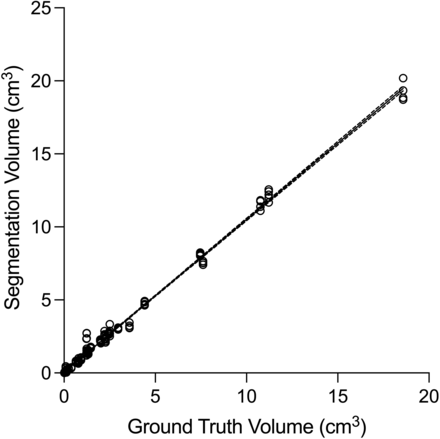
Linear regression of tumor volume measured by human operators versus ground truth volume showed excellent fit with R2 = 0.9951 (Fig 4). Bland-Altman analysis showed a bias of 0.14 cm3 toward larger tumor volumes with BLAST compared with the ground truth (95% limits of agreement, −0.38−0.66). There was excellent interreader agreement for tumor volumes with ICC = 0.9978 (95% CI, 0.9967−0.9987). The median segmentation time using BLAST for all operators was 2.8 (IQR, 1.6–4.3) minutes/metastasis and was not influenced by tumor size.
Comparison of tumor volumes measured by operators with BLAST versus ground truth. Linear regression of the data (dashed lines indicate 95% CI) shows an excellent fit (R2 = 0.9951). The measured volumes closely approximate the ground truth volumes, with a slope close to unity (1.05; 95% CI, 1.04–1.06).
Forty-five contours (94%) were scored a 4 or 5, indicating that the contours were acceptable clinically for treatment, requiring no changes at all or minor edits that are not thought to be clinically relevant. Only 3 contours (6%) scored a 3, indicating that minor edits were needed.
DISCUSSION
The treatment of brain metastases with SRS requires accurate detection and segmentation, which can be time-consuming and challenging. Furthermore, the follow-up of brain metastases necessitates reliable measurements of tumor burden to evaluate treatment response, which is important in routine clinical practice and clinical trials. To this end, methodologies for accurate and rapid segmentation of brain tumors have the potential to greatly impact the treatment of BM by improving the accuracy of treatments and measuring the response.
In the present work, we show the results of a methodology that provides highly accurate and reproducible segmentation of BM using multiparametric MR images. The methodology is based on using the statistics of normal background brain to identify abnormal tissue. As opposed to using K-means clustering to detect and segment BM directly, K-means clustering is used to identify normal background brain voxels from a section of normal brain, allowing them to be excluded and resulting in segmentation of enhancing tumor. Conveniently, the thresholds used to exclude nontumoral voxels are set relative to the statistics of the centroid of the background brain cluster.
Brain lesion segmentation based on the detection of outlier voxels has been previously described by Seghier et al.14 In their methodology, a fuzzy clustering procedure was used to identify outlier voxels corresponding to brain pathology from gray and white matter using T1-weighted images alone. The methodology was subsequently adapted to detect and segment BM on postgadolinium T1-weighted sequences, but the performance of the detection and segmentation task was limited by false-positives, mainly from vascular structures (arteries and veins), the dura, and the choroid plexus.15 These methods require training on a set of normal brains to model the intensity distribution of tissue types. One of the advantages of BLAST is that no training is required to perform segmentations because the normal brain cluster is defined on the basis of the statistics from the subject. Additionally, another main difference between these methods and BLAST is the use of multiparametric data in BLAST to better separate the normal brain cluster corresponding to gray and white matter from outlier voxels corresponding to BM and other normal structures such as blood vessels or the dura. In particular, the use of a black-blood sequence such as T2 FLAIR in combination with postcontrast T1 MPRAGE provides excellent separation of contrast-enhancing blood vessels from tumors in parameter space, resulting in fewer false-positives. This finding was highlighted in the results of the ablation study, which showed the superior performance of thresholding in both T1 MPRAGE and T2 FLAIR compared with thresholding in T1 MPRAGE alone.
The ablation study found no significant effect of background layer subtraction on the overall segmentation performance using BLAST. This finding is probably the result of adequate separation of enhancing tumor from the background layer in parameter space for most of the BM in this study, allowing the use of higher thresholds in T1 MPRAGE and T2 FLAIR. An analysis of the effect of background layer subtraction, however, showed that increasing levels of background layer subtraction improve segmentation performance at lower thresholds, which may be important for segmenting faintly enhancing metastases. Background layer subtraction also reduced the amount of background voxels included in the segmentation mask, possibly aiding users in the detection of metastases. Further work will be required to understand whether background layer subtraction provides any added benefit for segmenting or detecting faintly enhancing tumors or other pathologic processes whose signal characteristics overlap with normal brain (such as edema or nonenhancing tumor). Additionally, the optimal level of background subtraction for these applications will require further experimentation.
To evaluate the performance of segmentations using BLAST, we recruited human operators to use the methodology to generate contours of BM. Volumetric measurement of BM with the BLAST algorithm showed excellent interobserver agreement (ICC = 0.9978). Furthermore, tumor volumes measured with BLAST showed excellent correlation with ground truth manual segmentation. The excellent interobserver agreement and accuracy are features of BLAST that could make this algorithm well-suited for assessing response to treatment.
The results of the human operator study demonstrated a poorer segmentation performance for smaller metastases compared with larger metastases. Most interesting, this difference did not exist when evaluating BLAST performed by iterating a combination of thresholds in T1 MPRAGE and T2 FLAIR with background layer subtraction. The poorer performance of BLAST by human operators for smaller metastases was, therefore, seen to be a limitation of the users and the interface of the current Matlab implementation rather than a limitation of the BLAST algorithm. We expect that this situation will be improved in future versions of the software, which will allow operators to better visualize smaller metastases and allow real-time evaluation of the segmentation as thresholds are manipulated by the operator.
While a head-to-head comparison of BLAST and DL was not performed, compared with recently published state-of-the-art DL methods, BLAST fared well. Using a 2.5D fully convolutional neural network based on the GoogLeNet architecture (https://www.mathworks.com/help/deeplearning/ref/googlenet.html) trained on multiparametric MR imaging including 3D T1 BRAVO (GE Healthcare) postgadolinium, 3D T1 CUBE (GE Healthcare) pre- and postgadolinium, and T2 FLAIR, brain metastasis segmentation with a mean DSC = 0.79 (SD, 0.12) has been reported.3 With a 3D U-Net trained on 3D T1 echo-spoiled gradient echo postgadolinium or subtraction images, segmentation of BM with a median DSC = 0.75 and a median HD = 1.5 mm has been achieved.4 By means of a self-adaptive nnU-Net model (https://www.nature.com/articles/s41592-020-01008-z) trained on 3D T1 postgadolinium images, excellent segmentation performance with an overall mean DSC = 0.822 (SD, 0.095) for all metastases in the test set (mean size = 12.3 [SD, 9.2] mm) and DSC = 0.868 (SD, 0.075) for metastases of ≥6 mm has been recently shown.5 Further work will be required to evaluate the performance of BLAST compared with DL algorithms for brain tumor segmentation.
The median segmentation time for BLAST (2.8 minutes/metastasis) was longer compared with most DL algorithms, which report whole-brain inference times ranging from 20 seconds to 5 minutes.3,4,16,17 Inference times, however, may not be a fair comparison because they do not take into consideration the quality or clinical usability of the segmentations. A recently FDA-approved algorithm based on a 3D U-net and DeepMedic (https://github.com/deepmedic/deepmedic) of volumetric MR imaging and CT data showed whole-brain inference times of 90 seconds but an average of 6.1 minutes/case for users to finalize segmentations of BM for treatment planning.18
The median segmentation time presented herein is primarily a function of operator manipulation of thresholds and re-evaluation of the resulting segmentation, which varied between operators (IQR, 1.6–4.3) minutes/metastasis. With the current Matlab implementation, the process of adjusting the threshold requires a re-computation of the brain metastasis mask after each change in threshold, which can be a lengthy process. In the future, this time can be shortened with improvements in the user interface.
The clinical acceptance of the BLAST-generated contours measured using a 5-point Likert score was excellent. Of the 3 contours that required editing, 2 were small metastases that were located superficially in the brain, and partial volume averaging effects were the cause of the poor segmentations. For the other contour, an adjacent blood vessel altered the shape and size of the contour, requiring minimal editing.
There are several limitations to the presented technique:
Hemorrhagic metastases were excluded from the study. These hemorrhagic BM have a propensity for lower T2 FLAIR signal within areas of hemorrhage, which overlap in signal intensity with adjacent dura or blood vessels. Segmentation of these metastases could be aided in the future by additional parameters such as pregadolinium T1 MPRAGE, which can better distinguish between the brain metastasis and T2 hypointense normal brain structures such as the dura or blood vessels.
Another limitation of the algorithm is how the necrotic core is segmented. In the present study, the algorithm fills in the necrotic core on the basis of the boundaries of surrounding enhancing tumor. In some cases, areas of necrosis could be excluded if the rim of enhancing tumor is very poorly enhancing or thin. In the future, these areas of necrosis could be segmented separately with the addition of other parameters, including pregadolinium T1 MPRAGE.
The present study uses postgadolinium T2 FLAIR, which is not a widespread practice. At our center, T2 FLAIR is routinely acquired postgadolinium administration to save table time and enable a longer delay between gadolinium administration and the acquisition of T1 MPRAGE. The acquisition of T2 FLAIR after gadolinium administration also enables the detection of subtle leptomeningeal enhancement, which can be missed on T1 sequences alone.19 It is possible that the use of postgadolinium T2 FLAIR could provide an advantage for segmenting enhancing BM because the signal enhancement on T2 FLAIR resulting from intratumoral gadolinium leakage could result in better separation of background brain and the metastasis in T2 FLAIR parameter space. While this article has not addressed this possibility, our experience with BLAST has shown the feasibility of BLAST with pregadolinium T2 FLAIR data as well.
The current study is a single-center validation study with a small number of patients scanned using MR imaging systems from a single vendor. To address the possibility that the sample size was too small to detect a difference between arms, we calculated that a total sample size of 21 brain metastases would have been required to have an 80% power to assess the equivalence of the contoured volumes between the experimental method and the criterion standard, assuming an equivalence limit of ±20% of the criterion standard volume and an α threshold of .05, assuming that the dispersion of the results in the study reflected the true population dispersion. A larger sample size will be required in future studies to reduce the equivalence limit and increase overall power. The general utility of BLAST in a larger multicenter study of BM will also be needed to evaluate the generalizability of our results.
Despite these limitations, the segmentations produced by BLAST were easy to generate and could be used for BM treatment-planning or response-evaluation. The algorithm could also be adapted for diagnostic metastasis detection. The BLAST algorithm was able to detect and distinguish BM adjacent to arteries and other enhancing structures like venous sinuses and the dura, which can be challenging to detect. While it is possible that the BLAST algorithm alone may be sufficient for segmenting most BM, it can likely have the greatest impact through using the generated segmentations to train new DL models on improved and larger ground truth data sets for metastasis detection and segmentation.
Finally, the general framework of BLAST is not limited to metastases. The algorithm could be used to segment primary brain tumors (including enhancing and nonenhancing tumor subregions) or other brain pathology (such as white matter disease for instance). It could also be used with other MR imaging pulse sequences and in combination with other modalities, including CT and PET.
CONCLUSIONS
We present here an alternative methodology for brain metastasis segmentation, which provides accurate and reproducible segmentations of both parenchymal and dural metastases without extensive a priori training. Combined with the relative simplicity of the algorithm, the methodology could be widely implemented for brain metastasis treatment and response assessment.
Footnotes
C. Heyn is supported by a New Investigator Research Grant from the SickKids Foundation and the Canadian Institutes of Health Research.
C. Heyn, H. Soliman, and A.R. Moody are coinventors on a provisional patent assigned to Sunnybrook Research Institute (US Provisional Patent No. 63/537,092) for the methodology described in this article.
Disclosure forms provided by the authors are available with the full text and PDF of this article at www.ajnr.org.
Indicates open access to non-subscribers at www.ajnr.org
References
- Received May 24, 2023.
- Accepted after revision August 16, 2023.
- © 2023 by American Journal of Neuroradiology