Abstract
BACKGROUND AND PURPOSE: Differentiating glioblastoma from solitary brain metastasis preoperatively using conventional MR images is challenging. Deep learning models have shown promise in performing classification tasks. The diagnostic performance of a deep learning–based model in discriminating glioblastoma from solitary brain metastasis using preoperative conventional MR images was evaluated.
MATERIALS AND METHODS: Records of 598 patients with histologically confirmed glioblastoma or solitary brain metastasis at our institution between February 2006 and December 2017 were retrospectively reviewed. Preoperative contrast-enhanced T1WI and T2WI were preprocessed and roughly segmented with rectangular regions of interest. A deep neural network was trained and validated using MR images from 498 patients. The MR images of the remaining 100 were used as an internal test set. An additional 143 patients from another tertiary hospital were used as an external test set. The classifications of ResNet-50 and 2 neuroradiologists were compared for their accuracy, precision, recall, F1 score, and area under the curve.
RESULTS: The areas under the curve of ResNet-50 were 0.889 and 0.835 in the internal and external test sets, respectively. The area under the curve of neuroradiologists 1 and 2 were 0.889 and 0.768 in the internal test set and 0.857 and 0.708 in the external test set, respectively.
CONCLUSIONS: A deep learning–based model may be a supportive tool for preoperative discrimination between glioblastoma and solitary brain metastasis using conventional MR images.
ABBREVIATIONS:
- CE
- contrast enhanced
- GBM
- glioblastoma
- ROC
- receiver operating characteristic
- AUC
- area under the curve
Glioblastoma (GBM) and brain metastases are the most common malignant tumors in adults.1 These 2 entities have different treatment options, and it is therefore essential to distinguish them promptly to determine the proper treatment strategy. In patients with a history of underlying malignancy and conventional MR imaging findings of multiple enhancing lesions, a diagnosis can be made easily. However, approximately 25%–30% of brain metastases present as single lesions, and in lung cancer—the most common cancer to metastasize to the brain—approximately 50% of patients are thought to have brain metastases as the initial presentation.2,3 In addition, GBM and solitary brain metastasis have overlapping MR imaging features, including rim enhancement with perilesional T2 hyperintensity, and are thus difficult to differentiate preoperatively.4 However, GBM has an infiltrative growth pattern; therefore, tumor cells diffusely infiltrate beyond the enhancing portion, manifesting as a perilesional T2 hyperintense region. Brain metastases have similar MR imaging features; however, this perilesional T2 hyperintensity is primarily due to vasogenic edema caused by the leaky capillary vessels of the enhancing tumor.5,6 In an effort to detect these microstructural differences, various advanced MR imaging techniques, such as perfusion MR imaging, MR spectroscopy, and diffusion tensor imaging, have been applied to distinguish GBM from solitary brain metastasis, with particular emphasis on the aforementioned perilesional T2 hyperintense region.7⇓⇓-10 Collectively, these studies have shown promising results indicating that the perilesional T2 hyperintense region, along with the enhancing portion itself, carries valuable information that may preoperatively distinguish these 2 entities. However, advanced imaging techniques require additional scanning time, and their quantitative values can vary depending on the imaging parameters, posing difficult challenges for practical application.
Recently, radiomics have been used to analyze various textural and handcrafted features to classify or predict prognosis of disease through medical images that are beyond the perception of human eye.11,12 However, radiomics needs careful preprocessing steps, including delicate segmentation. Deep learning—a subfield in machine learning—extracts information directly from the data, omitting the step of manual feature extraction in decision making.13 In the field of neuro-oncology, specifically glioma imaging, previous studies have shown the potential of deep learning for classifying gliomas based on genetic mutations or clinical outcomes.14⇓-16
In this study, we hypothesized that deep learning may differentiate GBM from solitary brain metastasis without extraction of predefined features. Thus, we aimed to develop a deep learning–based model to differentiate GBM from solitary brain metastasis using preoperative T2-weighted and contrast-enhanced (CE) T1-weighted MR images and further validate its diagnostic performance.
MATERIALS AND METHODS
Patient Population
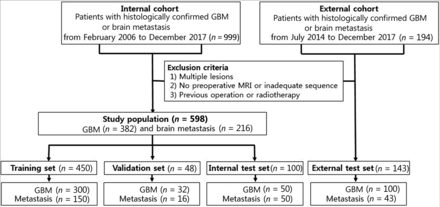
This retrospective study was approved by the institutional review board of our hospital, which waived the requirement to obtain informed patient consent. The records of 999 consecutive patients with histologically confirmed GBM or brain metastasis between February 2006 and December 2017 were retrospectively reviewed. Among these patients, those with preoperative MR images, including T2-weighted and CE T1WI, were included. Exclusion criteria included 1) multiple enhancing lesions; 2) patients with absent or inadequate MR images; and 3) patients with previous intracranial intervention, such as operation, gamma knife surgery, or radiation therapy. According to these criteria, 598 patients were included (357 men and 241 women; mean age, 57.4 ± 14.7 years). Fig 1 summarizes the study population selection.
Flow chart showing the patient population in the internal and external cohorts.
From the total study cohort, 450 patients were randomly selected for model training (300 GBM, 150 metastasis), and 48 patients (32 GBM, 16 metastases) were selected for model validation. The remaining 100 patients (50 GBM, 50 metastases) were left out on the patient level as an internal test dataset. The MR images of 143 patients (100 GBM, 43 metastases) at an outside tertiary referral hospital were used as an external test dataset; patients who satisfied the same inclusion and exclusion criteria as the internal cohort were extracted from their electronic database between January 2014 and December 2017.
MR Acquisition and Image Preprocessing
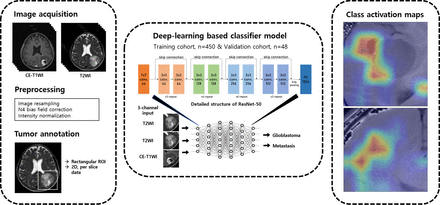
Preoperative imaging was performed using 1 of 4 3T MR imaging units (Ingenia or Achieva, Philips Healthcare; Discovery MR750, GE Healthcare; Tim Trio, Siemens) using an 8-channel sensitivity-encoding head coil. Details on the MR scanners and imaging parameters are summarized in the Online Supplemental Data. A diagram of the overall workflow is shown in Fig 2. The CE T1WI and T2WI of each patient were preprocessed, conducting intensity normalization by WhiteStripe normalization and N4 bias field correction. Images were resampled to 1 × 1 × 1 mm isotropic voxels. Preprocessed CE T1WI was coregistered to the T2WI. Rectangular-shaped ROIs were manually drawn on T2WI by a radiologist with 8 years of experience (B.S.) in MR imaging analysis using a conventional software package (MIPAV, National Institutes of Health) and confirmed by another radiologist with 13 years of experience (S.S.A.). ROIs were drawn on every section in which the mass was visualized on preoperative T2WI and included the peritumoral T2 hyperintense area, which was defined as a high signal intensity on T2WI beyond the border of the enhancing tumor portion.
Diagram of overall workflow.
Deep Learning Model
A 2D convolutional neural network (specifically the ResNet-50 model) with 50 layers consisting of 3-layer residual blocks17 pretrained with the ImageNet database was used. Hyperparameters of the fully connected layer of ResNet were fine-tuned using the training set data, and the convoluting and pooling layers were frozen. The batch size was 64, and a drop-out rate of 0.5 was applied with rectifier linear unit as the activation function. The model was trained for 100 epochs with stochastic gradient descent optimized with the Adam optimizer18 and the initial learning rate set to 0.001. Batch normalization was used in each layer to improve learning stability.19 Coregistered CE T1WI and T2WI were used as inputs to 2 of the 3 channels for training of our ResNet model. The same T2WI was inserted again into the last channel of our deep learning. Each section of CE T1WI and corresponding T2WI was treated as an independent image to increase the number of input data even though a group of slices belonged to the same patient. An ensemble learning method based on 5-fold cross-validation was used for model validation with most voting among models for final decision. Data splitting during training of the model was done per patient and not per section image to avoid overlapping bias. Regularization and fine tuning of hyperparameters of our model was done using the validation set (n = 48) from our institution. To establish the basis for judgment of our deep learning model, a class activation map was derived from each section of the images. All steps of the methodology were implemented with Python 3.7 and PyTorch v1.2 framework (https://pytorch.org/).
The final model was validated in an internal test set. The predictive index was defined as the number of slides classified as GBM by our classification model divided by the total number of slides per patient (ranging from 0 to 1). To determine the optimal cutoff value of the percentage of corrected slides for each patient, receiver operating characteristic (ROC) curves were derived. ROC curves were derived using SPSS version 25 (IBM).
Image Review by Neuroradiologists
The internal and external test datasets were reviewed by both experienced and junior neuroradiologists (S.S.A., neuroradiologist 1) and (I.S., neuroradiologist 2), with 13 and 4 years of experience, respectively. Both neuroradiologists were blinded to the pathologic and clinical information of all patients and were asked to classify each image as either GBM or metastasis, referring to the T2WI and CE T1WI. Subsequently, internal and external test sets were re-evaluated and classified again by both neuroradiologists, this time referring to the classification results of the ResNet-50 model.
Statistical Analysis
Patient demographics were compared between the GBM and metastasis subgroups using the independent 2-sample t test or chi-square test. The classification performance of the classification model and 2 neuroradiologists were evaluated on their accuracy, precision, recall, F1 score, and area under the curve (AUC). The 95% CIs of the precision, recall, and F1 scores were derived using the bootstrapping method with 1000 times 90% random sampling.
All statistical values were derived using SPSS version 25. The bootstrapping method was performed using R version 3.6.2 (http://www.r-project.org/). A P value ≤.05 was considered statistically significant.
RESULTS
Subjects
A total of 6617 axial slices of tumors from 598 patients with GBM or solitary metastases were included in the analysis. There was no significant difference in age and sex distribution between the GBM and metastasis groups in the internal and external cohorts; however, a higher percentage of patients had infratentorial lesions in the metastasis group (3.4% for the GBM group versus 22.7% for the metastasis group). Patients in the metastasis group included those with various primary tumor subtypes, most of which were lung cancer. The demographics of internal and external test sets are summarized in the Table.
Patient demographics of internal and external cohorts
Diagnostic Performance of Deep Learning–Based Model
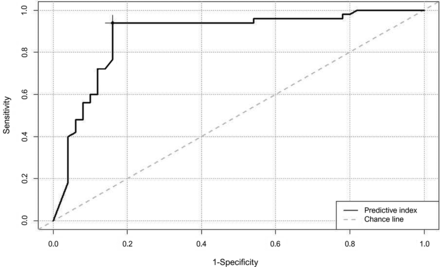
The optimal cutoff value for the predictive index was 0.55 when the AUC was 0.881 for the ROC curve drawn for the internal test cohort (Fig 3). The accuracy, precision, recall, F1 score, and AUC of the deep learning–based model were 89%, 0.852, 0.939, 0.893, and 0.889 in the internal test cohort and 85.9%, 0.907, 0.889, 0.893, and 0.835 in the external validation, respectively (Online Supplemental Data).
ROC curve for differentiating GBM and metastasis in the internal test set (AUC, 0.881; optimal cutoff for predictive index, 0.55).
In the internal test cohort, 8/50 (16.0%) metastases were misclassified as GBM, and 3/50 (6.0%) GBMs were misclassified as metastases. Similarly, 9/43 (20.9%) metastases were misclassified as GBM, and 12/100 (12.0%) GBMs were misclassified as metastases in the external test cohort. According to tumor location, 83 lesions were located in the supratentorial area, and 17 lesions were located in the infratentorial area in the internal test set. ResNet-50 miscategorized 10.8% (9/83) of the supratentorial lesions and 11.8% (2/17) of the infratentorial lesions. In the external test set, 130 lesions were supratentorial, and 13 lesions were infratentorial. All the infratentorial lesions were correctly categorized by ResNet-50, and all 21 miscategorized lesions were located in the supratentorial area in the external test set.
Because metastases were more prevalent in the cerebellum compared with GBM, our deep learning model seemed to recognize posterior fossa structures included in the ROIs, possibly contributing to the higher classification performance for infratentorial lesions (Figs 4 and 5).
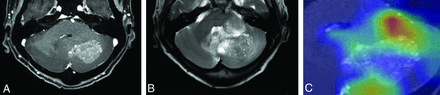
Images of a 65-year-old woman with history of recurrent ovarian cancer and pathologically proved brain metastasis. Contrast-enhanced T1WI (A) shows a heterogeneously enhancing mass in the left cerebellum. B, T2WI shows a perilesional T2 hyperintensity area surrounding the enhancing portion. C, Corresponding class activation maps show that the ResNet-50 model is referring to the mass and perilesional T2 hyperintensity area as well as the surrounding posterior fossa structures. ResNet-50 and both radiologists all correctly classified this lesion as brain metastasis.
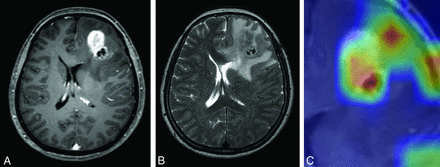
Images of a 62-year-old woman with pathologically proved GBM. CE T1WI (A) and T2WI (B) show an enhancing mass in the left frontal lobe with surrounding perilesional T2 hyperintensity area. Whereas ResNet-50 and radiologist 1 correctly classified this mass as GBM, radiologist 2 misclassified this lesion as metastasis. Corresponding class activation map (C) shows that ResNet-50 is referring to the enhancing portion as well as the surrounding peritumoral T2 hyperintensity areas.
Diagnostic Performance of Neuroradiologists
The accuracy, precision, recall, F1 score, and AUC of neuroradiologist 1 were 88.9%, 0.865, 0.918, 0.891, and 0.889 for the internal test set and 86.6%, 0.926, 0.88, 0.903, and 0.857 for the external test set, respectively. The accuracy, precision, recall, F1 score, and AUC of neuroradiologist 2 were 77%, 0.760, 0.776, 0.768, and 0.768 for the internal test set and 75.3%, 0.828, 0.82, 0.824, and 0.708 for the external test set, respectively (Online Supplemental Data).
Both neuroradiologists showed improved diagnostic performance after referring to the classification results of ResNet-50. The accuracy, precision, recall, F1 score, and AUC of neuroradiologist 1 after referring to ResNet-50 were 92.0%, 0.904, 0.940, 0.922, and 0.920 respectively, for the internal test set and 90.1%, 0.939, 0.920, 0.929, and 0.889 respectively, for the external test set. The accuracy, precision, recall, F1 score, and AUC of neuroradiologist 2 after referring to ResNet-50 were 91.0%, 0.940, 0.887, 0.913, and 0.910, respectively, in the internal test set and 88.0%, 0.895, 0.940, 0.917, and 0.839, respectively, in the external test set (Online Supplemental Data).
DISCUSSION
We proposed a deep learning–based model to differentiate solitary brain metastasis from GBM preoperatively using T2WI and CE T1-weighted conventional MR images. The model was developed using a large study population with varying scan parameters and validated in an external cohort and thus is expected to be robust and reproducible. Also, the classification model showed superior performance to that of the junior neuroradiologist and comparable results with those of the experienced neuroradiologist for both the internal and external test sets. Moreover, the classification model complemented the performance of the neuroradiologists, improving the classification performance of both junior and experienced neuroradiologists referring to the ResNet model.
It was noted that the deep learning–based model more frequently misclassified brain metastasis as GBM than GBM as metastasis. This might be because of the heterogeneity of the metastasis group, which included various primary cancer subtypes. In comparison, the GBM group included a histologically homogeneous group of patients. Although GBMs are known to have unique imaging and radiomics findings depending on their underlying genetic mutation statuses,20,21 they are thought to be relatively histologically homogeneous compared with metastases, which can have completely different histologic backgrounds.
In conventional deep neural networks, deeper networks are susceptible to the degradation problem, in which the network depth increases and the accuracy subsequently becomes saturated and degrades rapidly. ResNet-50 is a deep learning network that uses a residual learning framework that allows substantially deeper layers for training than that of conventional networks without degradation of performance. This type of deep learning network is capable of extracting more features and thus more accurately analyzing input images compared with conventional deep neural networks. Since its introduction, it has been widely used in various tasks in medical imaging, including detection, classification, and localization,22,23 showing comparable or better performance than that of conventional neural networks.24,25 Several imaging biomarkers have been studied to distinguish GBM from solitary brain metastasis. In previous studies, the shape of the enhancing portion, the signal intensity, and the extent of the peritumoral T2 hyperintensity were used to differentiate these 2 entities in conventional MR images.26,27 In those studies, nonspherical morphology of the enhancing portion and a higher normalized T2 signal intensity of the peritumoral portion were defining features of GBM. However, these previous studies had limitations in that they were conducted in small study populations.
Recent studies have applied radiomics-based machine learning methods to discriminate between GBM and solitary brain metastasis.28⇓–30 Radiomics-based machine learning models were used to preoperatively discriminate between these 2 entities based on CE T1WI. In that study, the best-performing supervised model showed accuracy of 85%.31 Another study used radiomics-based machine learning to distinguish between GBM and solitary brain metastasis.30 The researchers collectively investigated the diagnostic performances of 30 diagnostic models, with the 2 best-performing models showing an accuracy of 80%. Although these studies had somewhat promising results, they also had limitations in that the ROIs concentrated solely on the enhancing portions, failing to include data from the peritumoral portion and lacking external validation results. A recent study extracted radiomic features from an enhancing tumor portion and peritumoral T2 hyperintensity area of GBM and solitary brain metastasis and constructed a deep learning model based on these radiomic features.32 The study conducted external validation of the deep learning model with a high AUC value of 0.956. However, radiomics-based methods have innate limitations in that they include labor-intensive segmentation steps. To date, there is no study using an end-to-end deep learning–based method to discriminate between GBM and solitary brain metastasis. Our method had superior performance compared with those in the aforementioned studies using radiomics-based machine learning methods. Moreover, a strength of our deep learning model was that it could be robustly applied to conventional MR images with roughly drawn rectangular ROIs.
Our study had several limitations. First, instead of a 3D-based analysis, we used a 2D-based deep learning analysis to discriminate between GBM and solitary brain metastasis. However, considering that our training set was rather small (n = 450) for deep learning–based algorithm training, we reasoned that training with multiple MR image slices would be more suitable for adequate model training. Moreover, our deep learning model showed good performance in internal as well as external data, showing that our model has been properly trained. Second, because the patients had multiple MR slices, we arbitrarily adopted a new variable termed the “predictive index.” This variable required certain considerations because it was derived from a small internal test set; however, the diagnostic performance after adapting this cutoff in the external validation set demonstrated sustained discrimination performance. Third, in clinical practice, brain masses represent various clinical entities along with GBM and metastasis. These entities, such as lymphoma, demyelinating disease, infarction, and so on, should also be considered and integrated into the classification model in future studies. Finally, our model used only T2WI and CE T1WI, neglecting information from other sequences, such as T2 FLAIR images, and other advanced MR images, such as perfusion- or diffusion-weighted images. In addition, T2 FLAIR images are generally used to evaluate infiltrative nonenhancing glial tissue of GBM from vasogenic edema of brain metastasis. However, the heterogeneity of T2 FLAIR sequences (ie, precontrast versus postcontrast or 2D versus 3D acquisition) in our patient population prevented their use in training our deep learning model. Nevertheless, T2WI and CE T1WI are considered to be the most fundamental MR images and are almost always included in routine MR protocols, thus making our classification model more robust.
CONCLUSIONS
We developed a deep learning–based classification model to discriminate between GBM and solitary brain metastasis using conventional MR images. Our model had a diagnostic performance comparable with that of an experienced radiologist and had a complementary role in discriminating GBM and solitary brain metastasis. Therefore, deep learning may be used as an auxiliary tool for the discrimination of GBM from solitary brain metastasis.
Footnotes
I.Shin and H.Kim are co-first authors and contributed equally to this work.
Funded by Basic Science Research Program through the National Research Foundation of Korea, funded by Ministry of Science, Information, and Communication Technologies & Future Planning (2017R1D1A1B03030440)
Previously presented at: 105th Scientific Assembly and Annual Meeting of the Radiological Society of North America, December 1–6, 2019, Chicago, Illinois.
Disclosures: Sung Soo Ahn—RELATED: Grant: Basic Science Research Program through the National Research Foundation of Korea, Comments: 2017R1D1A1B03030440*. *Money paid to institution.
Indicates open access to non-subscribers at www.ajnr.org
References
- Received June 12, 2020.
- Accepted after revision November 13, 2020.
- © 2021 by American Journal of Neuroradiology