Abstract
BACKGROUND AND PURPOSE: In acute stroke patients with large vessel occlusions, it would be helpful to be able to predict the difference in the size and location of the final infarct based on the outcome of reperfusion therapy. Our aim was to demonstrate the value of deep learning–based tissue at risk and ischemic core estimation. We trained deep learning models using a baseline MR image in 3 multicenter trials.
MATERIALS AND METHODS: Patients with acute ischemic stroke from 3 multicenter trials were identified and grouped into minimal (≤20%), partial (20%-80%), and major (≥80%) reperfusion status based on 4- to 24-hour follow-up MR imaging if available or into unknown status if not. Attention-gated convolutional neural networks were trained with admission imaging as input and the final infarct as ground truth. We explored 3 approaches: 1) separate: train 2 independent models with patients with minimal and major reperfusion; 2) pretraining: develop a single model using patients with partial and unknown reperfusion, then fine-tune it to create 2 separate models for minimal and major reperfusion; and 3) thresholding: use the current clinical method relying on apparent diffusion coefficient and time-to-maximum of the residue function maps. Models were evaluated using area under the curve, the Dice score coefficient, and lesion volume difference.
RESULTS: Two hundred thirty-seven patients were included (minimal, major, partial, and unknown reperfusion: n = 52, 80, 57, and 48, respectively). The pretraining approach achieved the highest median Dice score coefficient (tissue at risk = 0.60, interquartile range, 0.43–0.70; core = 0.57, interquartile range, 0.30–0.69). This was higher than the separate approach (tissue at risk = 0.55; interquartile range, 0.41–0.69; P = .01; core = 0.49; interquartile range, 0.35–0.66; P = .04) or thresholding (tissue at risk = 0.56; interquartile range, 0.42–0.65; P = .008; core = 0.46; interquartile range, 0.16–0.54; P < .001).
CONCLUSIONS: Deep learning models with fine-tuning lead to better performance for predicting tissue at risk and ischemic core, outperforming conventional thresholding methods.
ABBREVIATIONS:
- AUC
- area under the curve
- DSC
- Dice score coefficient
- iCAS
- Imaging Collaterals in Acute Stroke
- IQR
- interquartile range
- Tmax
- time-to-maximum of the residue function
As demonstrated in recent Endovascular Therapy following Imaging Evaluation for Ischemic Stroke 3 (DEFUSE 3) and Extending the Time for Thrombolysis in Emergency Neurological Deficits (EXTEND) trials,1,2 perfusion imaging can be used to triage patients with acute ischemic stroke to reperfusion therapy in addition to the original “time window.” The DWI/PWI mismatch paradigm is the most common way of triaging patients,3 especially in those exceeding 6 hours of stroke onset.
The tissue at risk, sometimes called the penumbra, reflects the maximal extent of infarct if only minimal reperfusion is achieved, defined by time-to-maximum of the residue function (Tmax) > 6 seconds region using standard clinical software. Likewise, the ischemic core reflects the minimal ischemic lesion if major reperfusion is achieved, which has been defined by an ADC value < 620 × 10−6 mm2/s.4 Despite the simplicity and ease of use of single-value thresholds to identify salvageable tissue, such approaches have difficulty distinguishing benign hypoperfusion from tissue at risk5 and may fail to capture the complexity of the disease evolution.
Machine learning is a class of algorithms that automatically learn from data and provide predictions. Studies have shown that machine learning can be used to predict final stroke lesions from acute imaging data.6⇓⇓⇓⇓⇓⇓-13 Convolutional neural networks are a subtype of machine learning that do not require humans to define relevant features, instead extracting features automatically from images using many hidden layers (giving rise to the term “deep learning”).14⇓-16 One type of deep convolutional neural network known as a U-net has shown much promise for segmentation tasks in medical imaging.17
The most obvious approach to define the ischemic core and tissue at risk is to train 2 separate models using patients with complete or no reperfusion. However, such patients account only for a small subgroup of all patients who undergo reperfusion therapy, and the performance of deep learning models improves with increased sample size.18 Therefore, the aim of this study was to explore whether deep learning could provide a more accurate estimation of tissue at risk and ischemic core, and what is the most efficient and accurate approach with limited clinical data.
We evaluated 2 different approaches: training using targeted cases (patients with minimal and major reperfusion) only (separate training approach); or pretraining on a much wider cross-section of cases (including those with partial reperfusion) followed by fine-tuning on the targeted cases (pretraining approach). We hypothesized that the pretraining approach is superior to separate training and that both methods outperform the current clinical standard thresholding method based on the DWI/PWI mismatch.
MATERIALS AND METHODS
Patient Population
Patients with acute ischemic stroke were enrolled from 3 prospective, multicenter stroke trials: Imaging Collaterals in Acute Stroke (iCAS) from April 2014 to August 2017 (n = 128), DEFUSE from April 2001 to April 2005 (n = 74), and DEFUSE 2 from July 2008 to October 2011 (n = 140). iCAS19,20 is a multicenter observational study that enrolled patients with clinical acute ischemic stroke symptoms attributable to the anterior circulation, an NIHSS score of ≥ 5, and onset-to-imaging time of ≤24 hours. The DEFUSE and DEFUSE 2 protocols enrolled similar patients within a shorter time window (≤12 hours) and results have been reported.21,22
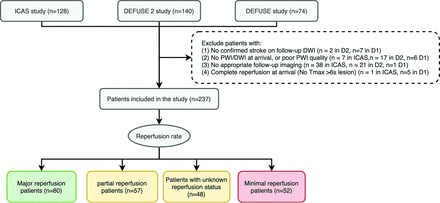
We excluded patients on the basis of the following criteria: 1) no confirmed ischemic stroke on follow-up DWI; 2) no PWI or DWI at arrival, or poor PWI quality; 3) no follow-up T2 FLAIR images within 3–7 days after stroke onset for iCAS and DEFUSE 2, or within 30 days for DEFUSE; or 4) complete reperfusion on initial PWI (no Tmax > 6 seconds lesion) (Fig 1).
Flow diagram of the study. According to the reperfusion rate calculated from baseline and 4- to 24- hour perfusion-weighted imaging, patients are grouped into major reperfusion (≥80%), partial reperfusion (20%–80%), minimal reperfusion (≤20%), and unknown reperfusion status (if 4- to 24-hour perfusion imaging was not performed). D1 indicates DEFUSE study; D2, DEFUSE 2 study.
iCAS (NCT02225730) and DEFUSE (NCT01349946) were approved by the institutional review boards of the participating institutions, and written consent was obtained for each participant. This study has been approved for retrospective analysis by the institutional review boards.
Imaging Protocol
All images were acquired at either 1.5T or 3T. Patients underwent MR imaging, including DWI (b=0 and b=1000 s/mm2) and dynamic susceptibility contrast-enhanced PWI using gadolinium-based contrast agents according to the standard protocol of each site. Postprocessing software (RAPID; iSchemaView) was used to reconstruct perfusion parameter maps: Tmax, MTT, CBV, and CBF. This software also automatically generates ADC segmentation with a threshold of <620 × 10−6 mm2/s and Tmax segmentation with a threshold of >6 seconds. Most patients underwent a follow-up PWI study within 24 hours, which was used to classify patients into minimal, partial, and major reperfusion as described below.
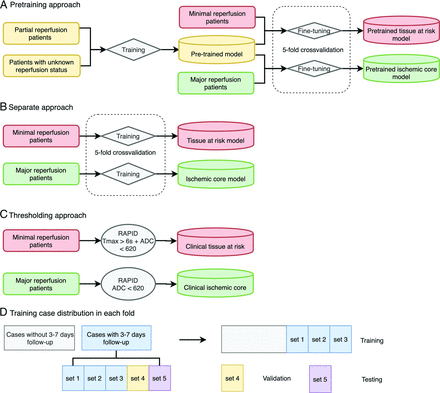
Patients with T2 FLAIR obtained at 3–7 days after stroke onset were used to evaluate the model performance; DEFUSE cases with DWI obtained at 4–8 hours and/or T2 FLAIR at 30 days after stroke treatment (because 24-hour and 3- to 7-day images were not part of the study protocol) were only used to train the deep learning algorithms but were not used for testing (Fig 2).
Illustration of 3 approaches to define tissue at risk and ischemic core and detailed case distribution during 5-fold cross-validation. The pretraining deep learning approach (A), the separate deep learning approach (B), and the thresholding approach (C). For the pretraining, 94 patients with partial/unknown reperfusion were used in the training set, and 11 patients were in the validation set. D, Cases without 3- to 7-day follow-up were used only in training (A and B), while patients with 3- to 7-day follow-up were used in training, validation, and testing. Patients with 3- to 7-day follow-up were divided randomly into 5 sets and used in training, validation, and testing with a ratio of 3:1:1. For each fold of the tissue-at-risk model, 37 cases were used for fine-tuning on the pretrained model, 6 for validation, and 6–7 for testing. For each fold of the ischemic core model, 48 cases were used for the fine-tuning; 13, for validation; and 13–14, for testing.
Imaging Analysis
Investigators at a core laboratory reviewed all studies. Neuroradiologists who were blinded to clinical information segmented the final infarct lesion on the follow-up studies. The segmented infarct lesions were used as ground truth for the deep learning model.
Patients were classified into 4 reperfusion categories based on the baseline and the 4- to 24-hour PWI study. We relied on the reperfusion rate rather than the TICI recanalization score to classify patients because it reflects the tissue reperfusion and predicts outcome better than recanalization.23,24 Reperfusion status was calculated as
Reperfusion Rate = 100% × (1 – [Tmax24hr > 6 seconds lesion / Tmaxbaseline > 6 seconds lesion]).
Patients with reperfusion rates of ≤20% and ≥80% were classified as having minimal and major reperfusion, respectively.25,26 Otherwise, they were classified as having partial reperfusion (if 4- to 24-hour PWI was available) or with unknown reperfusion (if not). Patients with minimal reperfusion were used to define tissue at risk, while those with major reperfusion were used to define ischemic core.
Imaging Preprocessing
All images were coregistered and normalized to the Montreal Neurological Institute template space using Matlab 2016b (MathWorks) and SPM 12 (http://www.fil.ion.ucl.ac.uk/spm/software/spm12). Of note, the spatial coverage of perfusion imaging was usually smaller than that of diffusion imaging, and only voxels with both diffusion and perfusion information were included in the model.
For input to the deep learning model, DWI (b=1000 s/mm2 images), ADC, Tmax, MTT, CBV, and CBF were normalized by the mean of their parenchymal tissue value. To preserve important information from the absolute value of Tmax and ADC, we created 2 masks separately for Tmax > 6 seconds and ADC < 620 × 10−6 mm2/s using simple thresholding.
Training Approaches
A neural network called attention-gated U-net was used in this study and was reported in previous literature12 (Online Supplemental Data and Online Fig 1). In short, the model takes 5 consecutive slices of DWI, ADC, Tmax, MTT, CBF, CBV, and masks of Tmax and ADC as input and gives a probability map of infarct segmentation with voxel values that ranged from 0 to 1 as output. A value close to 1 indicates that the voxel is more likely to be infarcted, while a value close to 0 indicates that the voxel is likely to be spared. The consecutive slices provided the model with more context than a single section of an image.
We explored the pretraining, separate, and thresholding approaches to test which one performed best (Fig 2). In the pretraining approach, a single model was first trained using patients with partial and unknown reperfusion status. Then, starting from these weights, 2 separate models were generated by fixing the weights in the encoder layers but fine-tuning the decoding layers, one using patients with minimal reperfusion to create a tissue-at-risk model and the other using patients with major reperfusion to create an ischemic core model. In the separate approach, 2 separate models were trained from scratch with patients with either minimal or major reperfusion. Because there were relatively fewer subjects who fell into these extreme cases, there was less data for each of the separate models for training. In the thresholding approach, the clinically used Tmax and ADC segmentations from RAPID were used. The union of Tmax > 6 seconds and ADC < 620 × 10−6 mm2/s was used to define tissue at risk. Tissue with ADC < 620 × 10−6 mm2/s was used to define the ischemic core.27
During the pretraining phase, 10% of the cases were used as a validation set and the rest were used for training. Five-fold cross-validation was performed for the separate approach and the fine-tuning part of the pretraining approach to reduce bias (Fig 2). Given the multicenter, multivendor nature of the dataset, this system represented the best test of the generalizability of the model.
Performance Evaluation
The area under the curve (AUC) was calculated for both the deep learning models and the Tmax and ADC thresholding method. The AUC was calculated for each case within the ipsilateral stroke hemisphere, except in 1 case for which there were bilateral strokes.
To calculate the Dice score coefficient (DSC) and lesion volume difference between prediction and ground truth, we set a threshold probability of .5 for all deep learning models. To calculate the mismatch ratio predicted by the models, we also applied the tissue-at-risk model to patients with major reperfusion, and the ischemic core model, to those with minimal reperfusion.
Statistical Analysis
Statistical analysis was performed using STATA (Version 15.0; StataCorp). The χ2 or Fisher exact test and the Kruskal-Wallis equality-of-populations rank test were performed for demographic and clinical information. Paired-sample Wilcoxon tests were performed to compare AUC, DSC, lesion volume difference, and absolute lesion volume differences between the pretraining approach and separate approach, as well as the pretraining approach and the Tmax/ADC thresholding methods. The concordance correlation coefficient (ρc) was used to analyze the lesion volume predictions. Because infarct volumes were not normally distributed, cubic root transformation was performed for the ρc calculation. The correlation was considered excellent with ρc > 0.70, moderate when ρc was between 0.50 and 0.70, and low with ρc < 0.50.28 All tests were 2-sided, and P ≤ .008 was considered statistically significant after adjustment by the Benjamini-Hochberg method.
RESULTS
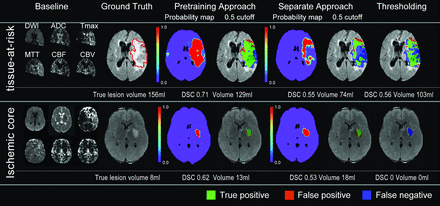
We reviewed 342 patients from DEFUSE 1, DEFUSE 2, and iCAS and eventually included 237 patients (Fig 1). Fifty-two patients were classified as having minimal reperfusion; 57, as partial reperfusion; 80, as major reperfusion; and 48, as unknown reperfusion. Clinical and imaging information is summarized in the Table. The time for training a model was 5 hours, and the time for generating prediction for each patient was 30 seconds with our current workstation. Figure 3 shows several examples of predictions using the 3 approaches. Online Supplemental Data show the effect of the attention map at each level in the U-net.
Two representative cases of predictions from the pretraining, separate, and thresholding approaches. Upper row: A 71 -year-old man treated with IV tPA only, which achieved 0% reperfusion. Of note, only voxels with both diffusion and perfusion information are included in the model. His true lesion is used to define tissue at risk. Lower row: A 45-year-old man treated with IV tPA and thrombectomy, which achieved 100% reperfusion. His true lesion is used to define ischemic core. The pretraining approach had more accurate prediction than either the separate approach or the thresholding method, both visually, with DSC analysis, and volumetrically (Online Supplemental Data). Green areas overlaid on the FLAIR image represent true-positive, blue represents false-negative, and red represents false-positive.
Clinical and imaging information of patients includeda
Prediction of Tissue at Risk
The evaluations were performed in 33 patients with minimal reperfusion with T2 FLAIR follow-up at 3–7 days. As shown in Online Supplemental Table 2, the pretraining approach achieved the highest AUC (0.92; interquartile range [IQR], 0.89–0.95) and DSC (0.60; IQR, 0.43–0.70) compared with the separate approach and thresholding method. There was no statistical difference in the volume difference or the absolute volume difference among the 3 approaches. However, the volume of tissue at risk predicted by the pretraining approach showed excellent concordance (ρc= 0.822; 95% CI, 0.725–0.919) with the true infarct volume compared with the separate (ρc = 0.685; 95% CI, 0.517–0.852) and thresholding (ρc = 0.657; 95% CI, 0.511–0.804) approaches. The volumetric agreement between the pretraining approach and the true lesion volume and the percentage volume difference are shown in the Online Supplemental Data and the Table.
Prediction of Ischemic Core
The evaluations were performed in 67 patients with major reperfusion with T2 FLAIR follow-up at 3–7 days. The pretraining approach again achieved the highest AUC (0.94; IQR, 0.89–0.97) and DSC (0.57; IQR, 0.30–0.69) compared with the separate approach and the thresholding method. The pretraining approach also showed less biased volume prediction compared with the thresholding approach and achieved excellent concordance (ρc= 0.756; 95% CI, 0.651–0.860) with the true infarct volume compared with the separate approach (ρc = 0.657; 95% CI, 0.519–0.795) and thresholding (ρc = 0.625; 95% CI, 0.489–0.762). The volumetric agreement between the pretraining approach and the true lesion volume and the percentage volume difference are shown in the Online Supplemental Data and the Table.
Mismatch Patterns Predicted from the Deep Learning Models
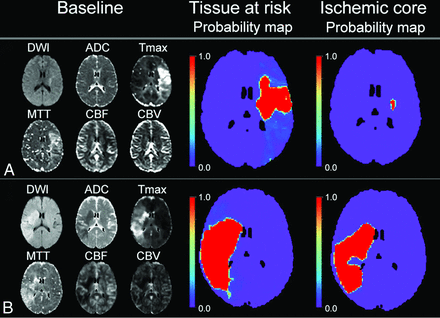
The median mismatch ratio yielded by the pretraining approach was 2.0 (IQR, 1.5–4.2) for patients with minimal reperfusion and 2.7 (IQR, 2.0–5.4) for patients with major reperfusion, compared with 2.9 (IQR, 1.4–6.8; P = .07) and 5.4 (IQR, 2.3–13.9; P < .001) given by the thresholding approach. Examples of mismatch predicted by the 2 different approaches are shown in Fig 4.
Example of how the deep learning models would be used for triage compared with the thresholding method. A, A 44-year-old woman with a large mismatch between tissue at risk and ischemic core prediction (mismatch ratio of 11.8), indicating a small stroke if the patient received successful treatment or a much larger stroke if the patient did not receive any treatment. B, 44-year-old woman with a small mismatch ratio of 1.4, indicating limited tissue salvage despite successful treatment. This illustrates how estimated tissue outcome (with and without reperfusion) can be obtained from the deep learning approach and can facilitate clinical decision-making on whether to treat the patient (see Online Supplemental Data for detailed image).
DISCUSSION
By analyzing data from 3 multicenter clinical trials, this study showed that a pretraining approach using deep learning in which a large heterogeneous population is used to first train a common model, which is then bifurcated into models for minimal and major reperfusion, performs better than using separate training in a smaller group of patients with extreme reperfusion. Furthermore, it outperformed current clinically available prediction methods based on a threshold of the DWI/PWI mismatch for identifying tissue at risk and ischemic core in acute ischemic stroke.
Currently, Tmax > 6 seconds and ADC < 620 × 10−6 mm2/s are the state-of-the-art estimations for tissue fate with no reperfusion and complete reperfusion.1,22,27 However, these thresholds are derived from linear analysis and have not been validated in large cohorts.29 Factors such as collateral status and gray/white matter content may result in different susceptibilities to ischemia.30 This study suggested that the single-valued thresholding approach could be outperformed using a nonlinear analysis method such as deep learning. While we have shown the capabilities of deep learning using MR imaging as the initial imaging study, we recognize that CT is becoming increasingly used for stroke triage. Similar methods could likely be used with CT data, and this MR imaging–based approach using pretraining could act as a starting point for training a CT-based triaging system, given the wide availablity of CT scanners and the ability to extract similar perfusion parameters.
Previous traditional and machine learning studies used only patients with minimal and major reperfusion to generate the criteria of tissue at risk and ischemic core.6,25,26 In patients with ischemic stroke who received reperfusion therapy using the latest devices, >50% of patients have partial reperfusion when the reperfusion rate at 24 hours is intermediate (20%–80%) or the TICI score is between 2a and 2b.1,31,32 Our results show that an approach that trained models separately in patients with minimal and major reperfusion had only moderate correlation with true lesion volume and had only a minor advantage compared with conventional thresholding methods. This finding is likely because the separate approach “wastes” many cases that could potentially be used to improve the network prediction. If we wanted to use the separate approach to achieve the same level of predictive accuracy as the pretraining approach, it likely requires a much larger training set, which is challenging for clinical studies. For example, in the current study, the pretrained model ultimately had access to approximately 2–3 times more individual cases for training than the separate models. Therefore, the pretraining approach (fine-tuning on a pretrained model) is a promising approach to maximally use all available stroke data to improve the performance.
Fine-tuning techniques have been discussed in previous literature.33 Fine-tuning on the last layer is preferred when the prediction task is within the pretrained model, while fine-tuning on the last several layers is preferred for a more specific task as in this study. Previous studies have shown that using models pretrained on nonmedical image data may also perform well in medical imaging data.34 However, medical images such as MR imaging and CT are often quantitative or semiquantitative and in gray-scale, differing from nonmedical photos. Complicated network structure, filters, and pre-extracted features for regular photos may be resource-consuming and redundant for MR imaging and CT data and may not offer much performance benefit.35 This study showed that pretraining using medical imaging data of the same category and same cohort achieved excellent performance. In the future, establishing pretrained models exclusively for medical imaging may help translate deep learning models into clinical workflow application most efficiently.
Compared with previous studies that used machine learning and deep learning to predict tissue fate,6,7,12 the accuracy and visual reliability of our model are promising. Some may argue that if more cases are used in the training set, they will always benefit the model performance. A previous study that trained prediction models in all patients with stroke regardless of reperfusion status 12 showed good accuracy but had biased prediction in patients with minimal and major reperfusion, with under- and overestimations in lesion size, respectively. The current study shows that refining the training strategy to specifically include patients with extreme reperfusion states as a fine-tuning step will provide less biased predictions. McKinley et al6 trained 2 random forest classifiers on 15 cases with TICI 3 (ischemic core classifier) and 10 cases with TICI 0 (penumbra classifier). They reported a mean DSC of 0.32 [SD, 0.23] in cases with TICI grades 1 and 2a and 0.34 [SD, 0.22] in cases with TICI grades 2b and 3. The models presented in this study appear to perform better, though it is difficult to compare metrics across studies that used different models and datasets. Therefore, validating our models in the same dataset is an important step for translation to clinical practice in the future.
After careful validation in a separate cohort and further improvement in predictive accuracy, the models can be applied to the triaging system in emergency departments. Similar to the current commercial software and workstations that apply 2 thresholds (Tmax > 6 seconds and ADC < 620 × 10−6 seconds/mm2), patients' images could be fed separately into the 2 models, which then generate predictions of tissue at risk and ischemic core. A larger mismatch ratio between tissue at risk and ischemic core indicates more benefit from reperfusion treatment, which can facilitate the timely clinical decision-making for patient triaging (Fig 4). However, new criteria for the cutoff of the mismatch ratio would be required because the current target mismatch criteria4,21 were established solely with the thresholding approach.
There are several limitations to this study. Treatment varied with respect to the use of thrombectomy and thrombolysis. Although we considered the most important factor, reperfusion status, in this study, clinical factors such as age, onset time to imaging, or other risk factors were not included in the analysis. It is our future aim to incorporate such clinical factors into the deep learning models and test whether it can further improve performance. The patient cohort in this study mainly had onset-to-imaging time exceeding 4.5 hours, which may affect the association between baseline imaging and tissue fate. However, no consensus has been reached on whether the perfusion profile is time-dependent,36,37 and patients presenting with prolonged symptoms represent the most important cohort for a clinical imaging triaging system. The model may not be directly applicable to images in the original space because the training data were in the Montreal Neurological Institute template space. However, the template space may help reduce the model overfitting and provide important spatial information. The model may be further fine-tuned with data in the original space to reduce the processing time in real clinical settings.
We did not perform outcome analysis because the dataset was not ideal for this purpose. Further studies are required to investigate whether using the model prediction improves clinical outcome, but it stands to reason that given the choice, a method that more accurately identifies dead and at-risk tissue would allow clinicians to make better decisions about thrombectomy. The data processing and model parameters were chosen on the basis of previous experience, and we did not extensively search all combinations of hyperparameters or fine-tuning techniques, given time and computational constraints. Although better combinations could provide improvement, our study demonstrated the feasibility of using pretraining for stroke imaging prediction and can be used as a jumping-off point for future studies seeking even better performance. Further studies are also warranted investigating whether prediction directly from source perfusion images will improve the performance.
CONCLUSIONS
This multicenter study showed that an attention-gated deep convolutional neural network can be used to identify tissue at risk and core in acute ischemic stroke at levels superior to the current clinical state of the art. Further clinical validation is required for these methods to be incorporated as a deep learning acute stroke triaging system.
ACKNOWLEDGMENTS
We appreciate the statistical consultation provided by Jarrett Rosenberg, PhD, and Tie Liang, EdD, Radiologic Sciences Laboratory, Department of Radiology, Stanford University.
Footnotes
This study was funded by National Institutes of Health (R01-NS066506, R01-NS039325).
The funders had no role in study design; collection, analysis, and interpretation of data; the writing of the report; and in the decision to submit the paper for publication.
Paper previously presented at: Annual Meeting of the American Society of Neuroradiology, May 19–23, 2019; Boston, Massachusetts; and International Stroke Conference, February 18–21, 2020; Los Angeles, California.
Disclosures: Yannan Yu—RELATED: Support for Travel to Meetings for the Study or Other Purposes: Stanford University; UNRELATED: Employment: Stanford University. Yuan Xie—UNRELATED: Employment: Subtle Medical Inc. Thoralf Thamm—RELATED: Grant: National Institutes of Health, Comments: Project No. 2R01NS066506-04A1.* Enhao Gong—UNRELATED: Board Membership: Subtle Medical; Employment: Subtle Medical; Stock/Stock Options: Subtle Medical. Søren Christensen—UNRELATED: Stock/Stock Options: iSchemaView, RAPID. Michael P. Marks—RELATED: Grant: National Institutes of Health*; UNRELATED: Board Membership: ThrombX Medical, Comments: no funds received but do have stock; Stock/Stock Options: ThrombX Medical. Maarten G. Lansberg—RELATED: Grant: National Institute of Neurological Disorders and Stroke.* Gregory W. Albers—UNRELATED: Consultancy: Genentech, iSchemaView; Stock/Stock Options: iSchemaView. Greg Zaharchuk—RELATED: Grant: National Institutes of Health*; UNRELATED: Board Membership: Subtle Medical; Expert Testimony: various Medico-legal consulting; Grants/Grants Pending: various National Institutes of Health projects, GE Healthcare, Bayer Healthcare*; Royalties: Cambridge University Press; Stock/Stock Options: equity, Subtle Medical. *Money paid to the institution.
References
- Received July 30, 2020.
- Accepted after revision December 28, 2020.
- © 2021 by American Journal of Neuroradiology