Abstract
BACKGROUND AND PURPOSE: Aberrant gyral folding is a key feature in the diagnosis of many cerebral malformations. However, in fetal life, it is particularly challenging to confidently diagnose aberrant folding because of the rapid spatiotemporal changes of gyral development. Currently, there is no resource to measure how an individual fetal brain compares with normal spatiotemporal variations. In this study, we assessed the potential for automatic analysis of early sulcal patterns to detect individual fetal brains with cerebral abnormalities.
MATERIALS AND METHODS: Triplane MR images were aligned to create a motion-corrected volume for each individual fetal brain, and cortical plate surfaces were extracted. Sulcal basins were automatically identified on the cortical plate surface and compared with a combined set generated from 9 normal fetal brain templates. Sulcal pattern similarities to the templates were quantified by using multivariate geometric features and intersulcal relationships for 14 normal fetal brains and 5 fetal brains that were proved to be abnormal on postnatal MR imaging. Results were compared with the gyrification index.
RESULTS: Significantly reduced sulcal pattern similarities to normal templates were found in all abnormal individual fetuses compared with normal fetuses (mean similarity [normal, abnormal], left: 0.818, 0.752; P < .001; right: 0.810, 0.753; P < .01). Altered location and depth patterns of sulcal basins were the primary distinguishing features. The gyrification index was not significantly different between the normal and abnormal groups.
CONCLUSIONS: Automated analysis of interrelated patterning of early primary sulci could outperform the traditional gyrification index and has the potential to quantitatively detect individual fetuses with emerging abnormal sulcal patterns.
ABBREVIATIONS:
- GI
- gyrification index
- GW
- gestational weeks
In the human cerebral cortex, primary sulci are important anatomic landmarks for predicting several primary, secondary, and higher order cortical areas.1 Their global patterning (arrangement, number, and size of primary sulcal folds) has been hypothesized to relate to the optimal organization of functional areas and their white matter connections.2⇓–4 Interestingly, the primary sulcal pattern is prenatally determined and appears to show little individual variability in healthy patients in early fetal life before 30 gestational weeks (GW).5⇓⇓–8 However, in many brain malformations and psychiatric/neurologic disorders, defects in the neurodevelopmental process result in a disrupted primary sulcal pattern, which may appear in early fetal life.9⇓⇓⇓–13 Therefore, sulcal folding analysis in the fetal brain may provide a useful tool for identifying the early signs of developmental brain disorders.
Previous studies using MR imaging have quantified cortical folding development in human fetal and preterm brains. The temporal changes of the gyrification index (GI) were observed during fetal life,14,15 and cortical surface curvatures at the global level have been measured to quantify the overall degree of cortical folding from 22–39 GW.15⇓⇓⇓⇓–20 The curvature and depth changes of cortical folding at the vertex level have also been observed for the entire cortical area.18,21,22 However, no quantitative analysis of the interrelated arrangement and global patterning of primary sulci in the human fetal brain exists. We previously developed a comprehensive and quantitative analytical method for comparing primary sulcal patterns by using not only the regional features of sulci folds themselves, but also the intersulcal geometric and topologic relationships.23 This method provided an effective means for detecting genetically influenced abnormal sulcal patterns in pediatric patients with cerebral malformations10,12 and developmental dyslexia.13
We hypothesized that the global pattern of primary sulci may be disrupted early in fetal brain development in fetuses destined to have abnormal sulcation at birth. Therefore, by using normal fetal brain templates to define normal sulcal patterning, we set out to measure early differences of sulcal pattern in fetuses with brain abnormalities confirmed by MR imaging at birth. The biologic motivation behind this hypothesis is that cortical arealization and connectivity begin early in fetal brain development and, if defective, might give rise to atypical sulcal topology. We also compared our method with an established metric, the GI, which has been used to quantify the development of human cortical folding.18,24
Materials and Methods
Patients
For this pilot feasibility study, fetuses with confirmed brain abnormalities were identified retrospectively from the existing clinical MR imaging data at Boston Children's Hospital. We searched for fetal MRIs performed between 18 and 30 GW with reports raising a concern for cerebral malformations and included only those fetuses who had the cerebral malformation confirmed by a follow-up postnatal brain MR imaging. The clinical imaging diagnoses were confirmed by 2 pediatric neuroradiologists (E.Y. and P.E.G.). Among approximately 40 fetuses identified by using this process, most were excluded because of serious head motion in the fetal MRIs, which could not be sufficiently processed to yield accurate fetal brain surfaces. Thus, 5 fetuses with confirmed brain malformations were included in the final analysis (male/female, 3/2; age [mean ± SD], 20–30 GW [25.3 ± 3.8]; maternal age [mean ± SD], 29–37 years [33.8 ± 2.7]) (1 Chiari II malformation [subject ID: M1] with sacral myelomeningocele, 3 polymicrogyria [M2, M3, and M5], and 1 atypical gyral folding [M4]). Fetuses M1 and M2 were initially thought to have a normal sulcal folding, but neonatal MR imaging identified atypical sulcal folding and polymicrogyria, respectively. One patient with polymicrogyria was longitudinally scanned with fetal MR imaging at 22 and 29 GW [M5a and M5b].
Healthy control patients were recruited from mothers referred to the Advanced Fetal Care Center at Boston Children's Hospital because of a family history of congenital heart disease, but for whom fetal echocardiogram was normal. Inclusion criteria were the following: maternal age of 18–45 years and gestational age of 18–30 GW. We excluded multiple gestation pregnancies and fetuses with dysmorphic features on sonography, brain malformations, other brain lesions, known chromosomal abnormalities, other identified organ anomalies, and known congenital infections. We also excluded patients as healthy controls if the fetal MR imaging identified these conditions as being present. Using these criteria, 21 healthy control fetuses were recruited and scanned, but 7 fetuses were excluded because of serious head motion. Fourteen healthy fetuses were finally included in this study (male/female, 4/10; age [mean ± SD], 21–30 GW [25.5 ± 2.3]; maternal age [mean ± SD], 25–39 years [29.9 ± 4.0]).
This study was approved by the institutional review board committee of Boston Children's Hospital.
MR Imaging Acquisition and Processing and Reconstruction of Cortical Plate Surface
Fetal brain MR imaging was performed by using a T2-weighted HASTE MR imaging sequence on a 3T scanner (Skyra; Siemens, Erlangen, Germany) equipped with the body matrix array (18 coil elements used) in combination with the spine array (12–18 coil elements) for a total of 30–36 coil elements. The following sequence was used for each patient: TR, ∼1.5 seconds; TE, 120 ms; FOV, 256 mm; in-plane resolution, 1 mm; section thickness, 2–4 mm. The HASTE acquisition was performed at least 3 times in different orthogonal orientations with respect to the fetal brain for reliable image processing.
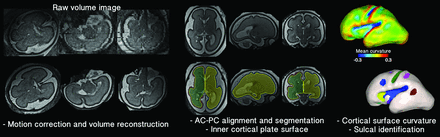
Fetal head motion was corrected, and 0.75 mm isotropic volume images were reconstructed from the multiple scans.25 The volume images were manually aligned along the anterior/posterior commissure points by using AFNI (http://afni.nimh.nih.gov/afni),26 and the cortical plate was semi-automatically segmented by using FreeView (http://surfer.nmr.mgh.harvard.edu). The cortical plate was painted on each section by using intensity value ranges and corrected via examination on orthogonal views. To reconstruct smooth surface models, the inner volume of the cortical plate was binarized and smoothed by using a 3 × 3 × 3 mean filter. The 3D inner cortical plate surface was then reconstructed by using the isosurface function with the isovalue of 0.5 in Matlab software (MathWorks, Natick, Massachusetts) (Fig 1). Although small folds similar in size to a voxel might be eliminated, our smoothing was effective to avoid noisy features and voxelized representation of the surface.
Processing overview for the identification of sulcal basins on the cortical plate surface. A raw volume image of an individual fetal brain (26 GW) is reconstructed to a motion-corrected, high-resolution volume. The reconstructed volume image is aligned along the anterior/posterior commissure (AC–PC) points. The inner volume of the cortical plate is semi-automatically segmented, its 3D surface is reconstructed, and its mean curvature is represented by a color map. Sulcal catchment basins are identified on the cortical plate surface based on the curvature and identified with different colors.
We used 9 previously published fetal brain templates from 23–31 GW that were constructed from 80 normal fetuses (http://brain-development.org/brain-atlases)27 as a reference for our sulcal pattern analysis. The cortical plate surfaces of the fetal templates were reconstructed by using the same process (Fig 2).
Sulcal matching overview. A, The combined set of sulcal folds (top) identified and generated from 9 fetal brain templates (bottom). Sulcal basins and their average positions are represented by black line and sphere, respectively. B, Sulcal matching result for an individual fetal brain (26 GW). Each sulcal basin of the individual brain is optimally matched with 1 of the sulcal basins of the 9 template brains. The spheres with the same color represent the matched corresponding sulcal basins between the templates and the individual brain.
Identification and Geometric Measurements of Sulcal Catchment Basins
Early sulcal catchment basins were used for sulcal pattern analysis because they are relatively invariant between patients and hypothesized to relate to functional areas and activations.28,29 Sulcal basins are concave substructures decomposed from one large part of cortical folds.29 Cortical mean curvature and depth on a surface model were measured by using FreeSurfer (http://surfer.nmr.mgh.harvard.edu), and sulcal basins were automatically identified by using a watershed algorithm based on the curvature map12,13,23,29 (Fig 1). A minimum bounding box for a given surface model was defined, and an average 3D relative position (x, left–right [0–1]; y, posterior–anterior [0–1]; z, inferior–superior [0–1]) was calculated for each sulcal basin. We also computed normalized surface area (s) and mean sulcal depth (d) for sulcal geometry.
Sulcal Pattern Matching and Similarity to the Templates and Automatic Sulcal Labeling
Sulcal pattern was represented with a feature set of sulcal basins including not only the local sulcal features, but also the intersulcal geometric relationships in the left and right hemispheres.23 To quantify the deviation from the normal pattern of sulcal folding for each fetus, individual brains were matched and compared with the combined set of all 9 fetal brain templates (Fig 2). Sulcal sets P (individual brain) and Q (9 template brains) containing m and n sulcal basins, respectively, were represented as follows:
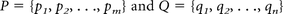
For the comparison, we used a spectral matching technique to determine the optimal match having the minimum difference of the features between 2 sulcal sets.13,23,30 We constructed a matrix M to store the affinities of all candidate matches and chose a subset matrix A, determining the optimal match and maximum affinity between P and Q by calculating the principal eigenvector of M. For a more detailed explanation of the methodologic procedures, see Im et al.23 Through this process, each sulcal basin of an individual brain was optimally matched with 1 of the sulcal basins of the 9 template brains (Fig 2), and the similarity to the templates was calculated, which ranged from 0–123,30 (On-line Fig 1). For the similarity measure, we used variable weighting of features to assess their relative importance on sulcal pattern similarity. First, all 3 features were optimally weighted in the sulcal pattern matching and similarity measure (wx,y,z = 3; ws = 8; wd = 0.8). Then, we evaluated the impact of each individual feature on similarity measures by setting all weights of the other features to 0.
Because the 3D position of the sulcal basin was required for optimal sulcal matching, we aligned the orientation of the cortical surface along the anterior/posterior commissure line. Although this is not a perfect spatial normalization, it was considered sufficient for measuring the scale-free value of relative position. Moreover, our method is insensitive to the spatial normalization because the intrinsic sulcal pattern characterized by intersulcal relationships is used for computing a large part of the sulcal pattern similarity and minimally affected by the spatial alignment.23
3D Global Gyrification Index
A GI is defined as the ratio between the whole areas of the cortical surface and its convex hull in 3D.14,31 First, the inner volume of the cortical plate surface was isolated and constructed, and the 3D morphologic closing operation was performed by using a sphere of 15 mm diameter as the structural element to close the sulcal folding.31 The outer hull surface wrapping the cortical plate surface was created from the binary closed volume by using the isosurface function (On-line Fig 2). We then calculated the 3D global GI of the left and right whole cerebral hemispheres.
Statistical Group and Individual Analysis
Using the sulcal pattern matching and comparison method, the sulcal pattern similarities to the set of 9 normal templates were computed with the 4 different feature sets for each fetus in the left and right hemispheres (whole feature, 3D position, sulcal area, and sulcal depth). The similarities to the templates were statistically compared between normal fetuses and fetuses with brain abnormalities with an independent 2-sample t test. The Pearson correlation and Mann-Whitney U test were used in normal fetuses to test the effects of gestational age and sex, respectively, on the sulcal pattern similarity. The group difference in GI was statistically assessed by using linear regression analysis. The dependent variable was the GI, and the independent variable was the group. Age and sex were used as covariates to control for their effects. Because 1 patient with an abormal brain had repeated longitudinal measurements (M5a and M5b), the group comparisons in the sulcal pattern similarity and GI were performed 2 times, including either M5a or M5b, to comply with the assumption of statistical tests.
We examined if the similarity of each individual abnormal brain to the templates deviated from the normal range by measuring its relative position in the distribution of all 14 normal individual fetuses. We counted the number of normal patients who had a lower similarity and expressed that value as a ratio to the total number of normal individual patients (histogram ranking). If an individual abnormal fetus showed a ratio of 0, meaning none of the normal individual fetuses had a lower similarity to the normal template, that patient was tagged as presenting an “abnormal” pattern of sulcal folding. If the ratio value was smaller than 0.1, the sulcal folding was deemed “suspicious” for an abnormal sulcal pattern.
Results
Sulcal Pattern Similarity to the Normal Templates
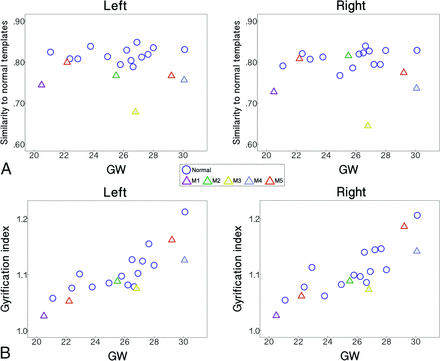
The fetuses with abnormal brains showed significantly lower sulcal pattern similarities to the templates compared with the normal fetuses in both hemispheres measured with the whole set of features (Fig 3A). In the statistical tests for each individual feature, significantly low similarities were observed in the abnormal group for the relative position, area, and depth of cortical folding in the left hemisphere. In the right hemisphere, sulcal position and depth pattern similarities were significantly lower in the abnormal fetal brains. All statistical results for the group comparisons with M5a or M5b are presented in Table 1. We confirmed that sulcal pattern similarities were not significantly associated with GW (left: correlation coefficient r = 0.075, P = .80; right: r = 0.334, P = .24) and not different between male and female patients (left: P = .37; right: P = .84).
Scatterplots of the similarity to normal templates computed with A, the whole feature set and B, GI against GW for the left and right hemispheres.
Statistical results for the comparisons of the sulcal pattern similarity with the templates between normal and abnormal groups with different feature sets
Regression analysis showed that global GI of the fetal brains increased with GW (statistical results including M5a [a] and M5b [b], left and right: P < .001), but there was no significant sex effect ([a] left: P = .38, right: P = .67; [b] left: P = .45, right: P = .78). The GI was not statistically different between the normal and abnormal fetal groups in either hemisphere ([a] left: P = .18, right: P = .22; [b] left: P = .33, right: P = .48) (Fig 3B).
Individual analysis based on the ratio quantifying the deviation from the normal showed that all abnormal fetal brains had an abnormal sulcal pattern with the ratio 0 in 1 or more feature sets in at least 1 hemisphere (left: M1, M2, M3, M4, and M5b determined as abnormal; right: M1, M3, M4, M5a, and M5b determined as abnormal). The similarity measures for all individual hemispheres and the ratio values for each abnormal fetal brain hemisphere are shown, along with 5 normal fetal brains for example, in On-line Fig 3. Our quantitative assessment for each abnormal fetal brain was compared with the qualitative assessment of fetal MR imaging. An abnormal sulcal pattern was detected for individual fetal brains that were judged to have abnormal gyrification by qualitative assessment (M3, M4, and M5). Moreover, high deviation from the normal was also quantitatively observed for fetal brains that were judged to have normal gyrification by qualitative fetal MR imaging assessment (M1 and M2). Demographic data and qualitative and quantitative assessments of fetal/neonatal MRIs are shown in Table 2. Specifically, abnormal brain M2, initially misjudged to have normal sulcal folding in qualitative fetal MR imaging but postnatally confirmed to have abnormal folding (false-negative), was assessed to be abnormal in left hemispheric sulcal pattern in our quantitative analysis (true-positive).
Demographic data, qualitative assessments of fetal and neonatal MRIs, and quantitative assessments of fetal MRIs for fetuses with brain abnormalities
Discussion
We used a novel automatic sulcal pattern comparison method to quantify sulcal pattern similarities of individual fetal brains against normal fetal brain templates by using multivariate geometric sulcal features and detected not only group differences between abnormal and normal fetal brains, but also individual differences in sulcal patterns in all abnormal fetuses. Although the entire set of features (3D position, area, and depth of sulcal basin) showed the highest statistical significance for the group comparison, the 3D position and depth of the sulcal basin were the major driving factors. The common cortical folding measurement GI was also used for the group analysis, but showed no significant difference between the groups. In our pilot data, our approach, based on sulcal location and depth patterns, outperformed the more traditional GI, previously thought to be the most sensitive measure for evaluation of the developing human brain.24 This suggests that in brain abnormalities associated with abnormal sulcal folding, the relative sulcal locations and depths are altered more than the overall amount of cortical folding during early fetal brain development.
Atypical patterns of sulcal locations and folding depths in fetal brains may indicate defective regulation of spatiotemporal dynamics of early cortical surface expansion and folding. Cortical areas do not develop independently, but in strong relation to other functional areas with optimized white matter connections, and accordingly show specific locations and size.3,32 It has been proposed that cortical functional arealization and organization may be related to the specific spatial pattern of early primary sulci in fetal brains.1,4 Similarly, the gyrogenesis theory suggests that areas of rapid growth form gyri at the center of a functional zone and underlie the formation of the first major folds during the early stage of cortical growth.33,34 The areal expansion and folding of the human cerebral cortex are precisely regulated processes in time during fetal life.5⇓⇓–8 The normal stereotypical pattern of gyrogenesis likely results in regular intersulcal relationships and folding depth at each fetal stage. In fact, the positional identity of cortical functional regions is defined by the combinatorial expression pattern of various genes, with their areal expansion also under tight genetic control with distinct spatiotemporal characteristics.32,35⇓–37 Therefore, atypical patterns of sulcal locations and folding depths in fetal brains before 30 GW likely are associated with defects in genetic control of cortical arealization and expansion.
In 2 cases (M1 and M2), our individual quantitative analysis detected an abnormal sulcal pattern not detected on qualitative fetal MR imaging assessment yet confirmed postnatally. Thus, our quantitative analysis shows the feasibility and potential for detecting emerging subtle abnormalities in sulcal patterns that are difficult to perceive visually. Future studies will be aimed at obtaining true sensitivity and specificity data in a larger cohort that includes postnatal imaging of normal fetuses because this pilot study could not show the full distribution of typical sulcal patterns because of the small number of normal fetuses. In addition, we will explore the ability of such quantitative analyses to decrease variation in clinical interpretations. Finally, comparison of quantitative assessments with genetic studies and follow-up clinical/behavioral outcomes is needed to further evaluate the potential applicability of our method in understanding the ability to detect and distinguish different genetic disorders and predict outcomes.
Limitations of MR imaging–based sulcal analysis on fetal brains include the low success rate (5/40 [12.5%]) for triplane reconstruction in clinically acquired fetal datasets and the current need for manual segmentations. However, because healthy control fetuses were scanned with a research MR imaging protocol that acquired more sets of triplane images, the success rate for our healthy fetal MR imaging processing was higher (14/21 [66.7%]). Future accelerations in image acquisition and improvements in postprocessing techniques are likely to increase success rates moving forward. Another limitation is the delay in the detection of cerebral sulci in MR imaging studies compared with histopathology. At least 1-week discrepancy has been reported between anatomic and qualitative MR imaging studies because of limitations in the resolution and contrast of in utero fetal MR imaging.38,39 If cerebral sulci are identified by using automatic and quantitative image processing techniques, sulcal detection is more delayed. For example, 1 small sulcus, the olfactory sulcus, was reported to appear before 20 weeks,7,8 but here it was not identified at such an early stage not only on the templates, but also on individual fetal brains. Delayed sulcal detection on fetal MR imaging has also been shown in previous quantitative studies.21,40 Ongoing innovations in this area as mentioned above are also likely to improve early sulcal detection.
Conclusions
Our quantitative sulcal pattern analysis with a focus on relative spatial location and depth of sulci has the potential to detect not only group differences in cortical folding, but also individual differences in fetuses destined to develop cortical malformations. Our method outperformed GI, which was dominated by gestational age. In addition, our method outperformed blinded expert qualitative fetal MR imaging assessments.
Footnotes
Disclosures: Kiho Im—RELATED: Grant: National Institutes of Health, Boston Children's Hospital. Caitlin Rollins—RELATED: Grant: National Institutes of Health, Comments: Scholar Award from the Pediatric Heart Network and K12 training award*; UNRELATED: Stock/Stock Options: Biogen. Cynthia Ortinau—RELATED: Mend A Heart Foundation; UNRELATED: Payment for Lectures (including service on Speakers Bureaus): Academy of Neonatal Nursing. Edward Yang—RELATED: Grant: National Institutes of Health, Comments: principal investigator for National Institutes of Health grant 1R21HD083956–01A1*; UNRELATED: Consultancy: Corticometrics LLC; Grants/Grants Pending: National Institutes of Health*. *Money paid to the institution.
This work was supported by National Institutes of Health grants 1R21HD083956-01A1, 1R01EB017337-01, and 1U01 HD087211-01; a Boston Children's Hospital Faculty Career Development Award; a Scholar Award from the Pediatric Heart Network supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health (U10HL068270); a Neurological Sciences Academic Developmental Award from the National Institutes of Neurological Disorders and Stroke; and the Mend a Heart Foundation.
Indicates open access to non-subscribers at www.ajnr.org
References
- Received October 21, 2016.
- Accepted after revision March 13, 2017.
- © 2017 by American Journal of Neuroradiology