Abstract
BACKGROUND AND PURPOSE: Fetal brain MR imaging is clinically used to characterize fetal brain abnormalities. Recently, algorithms have been proposed to reconstruct high-resolution 3D fetal brain volumes from 2D slices. By means of these reconstructions, convolutional neural networks have been developed for automatic image segmentation to avoid labor-intensive manual annotations, usually trained on data of normal fetal brains. Herein, we tested the performance of an algorithm specifically developed for segmentation of abnormal fetal brains.
MATERIALS AND METHODS: This was a single-center retrospective study on MR images of 16 fetuses with severe CNS anomalies (gestation, 21–39 weeks). T2-weighted 2D slices were converted to 3D volumes using a super-resolution reconstruction algorithm. The acquired volumetric data were then processed by a novel convolutional neural network to perform segmentations of white matter and the ventricular system and cerebellum. These were compared with manual segmentation using the Dice coefficient, Hausdorff distance (95th percentile), and volume difference. Using interquartile ranges, we identified outliers of these metrics and further analyzed them in detail.
RESULTS: The mean Dice coefficient was 96.2%, 93.7%, and 94.7% for white matter and the ventricular system and cerebellum, respectively. The Hausdorff distance was 1.1, 2.3, and 1.6 mm, respectively. The volume difference was 1.6, 1.4, and 0.3 mL, respectively. Of the 126 measurements, there were 16 outliers among 5 fetuses, discussed on a case-by-case basis.
CONCLUSIONS: Our novel segmentation algorithm obtained excellent results on MR images of fetuses with severe brain abnormalities. Analysis of the outliers shows the need to include pathologies underrepresented in the current data set. Quality control to prevent occasional errors is still needed.
ABBREVIATIONS:
- CNN
- convolutional neural network
- DC
- Dice coefficient
- IQR
- interquartile range
MR imaging of the fetal brain is an important adjunct to ultrasound in the detection and characterization of abnormalities at an early stage of development.1 High-resolution imaging is essential to accurately diagnose and follow up the evolution of these pathologies. New techniques have allowed the development of isotropic motion-corrected volume reconstructions based on the acquired 2D image stacks, including the so-called super-resolution reconstruction method.2 These types of volumetric reconstruction methods combine several stacks of 2D slices in different planes to construct a single isotropic volume, removing individual section artifacts and interslice inconsistencies,2⇓⇓⇓-6 as well as providing a volume with the same high resolution between slices as within a section. The segmentation of different parts and tissue types of the fetal brain should provide more accurate and more reproducible information regarding the evolution in certain pathologies, such as ventriculomegaly, malformations of cortical development, and tumors.
Performing such segmentation manually requires a high level of expertise and is time-consuming and prone to human error and variability; therefore, accurate automatic segmentation is essential for routine clinical use. Segmentation of the fetal brain is challenging because of the complex and rapidly changing anatomy during fetal life and is further complicated by variable image quality and a variety of artifacts.7
In medical image analysis, deep learning methods have recently proved to be very competitive, often outperforming conventional machine learning and model-based methods,8 including MR imaging of the adult and normal perinatal brain.9,10 Deep neural networks have the major advantage of being able to retrieve specific features for the task at hand directly from the data. The networks learn to extract and interpret features related to the segmentation task without the need to first derive a collection of handcrafted features from the image as input to a classifier or model. The state-of-the-art deep neural networks for segmentation are based on convolutional neural networks (CNNs). The use of automatic segmentation of fetal brain tissues by CNNs has been shown to be effective in normal cases11 and more recently in cases with spinal dysraphism as well for the ventricles.12 Developing automated segmentation tools for normal brains may be a good starting point, but in clinical practice, MR imaging is used to assess fetuses with pathology, rather than as a screening tool.13 Currently, there is a need for robust methods to segment fetal brain structures in the presence of varying severe abnormalities, which are common in the fetal period and can substantially affect the performance of developed techniques.10
One of the downsides of using CNNs is that they require large sets of training data.11 These data also have to be diverse enough for the CNN to be robust to pathologies. Typically, CNNs are trained by using empirical risk minimization to maximize the average segmentation performance. This can cause errors when pathologies are underrepresented in the training data set, as is typically the case with the available abundance of healthy control cases in contrast to the ones with abnormal findings. To address this problem, we specifically developed an algorithm that is more robust to anatomic abnormalities. This algorithm trains a CNN with distributionally robust optimization,14⇓⇓-17 which automatically reweighs the training samples with lower performance, encouraging the CNN to perform more consistently on all cases. This method has been shown to be more robust than conventional CNNs trained with empirical risk minimization.18 This algorithm was validated earlier using 197 fetal brain volumes from 4 different centers, including both normal brains and those with various CNS abnormalities. Using data from multiple centers with MR imaging machines from different vendors with various CNS abnormalities present during testing ensures us that this method of training is especially robust to different data input. This algorithm has since been updated to segment even more brain structures and was validated on a larger data set.19 The robustness of the algorithm allows the user to input a variety of new cases with abnormal findings with excellent overall generalization results, as is key for the clinical implementation of the algorithm. The acquired segmentations aid in the detection and characterization of fetal pathologies, which could be considered the most important goal of these automatic segmentations.10
The algorithm we use automatically segments the cerebellum, ventricular system, and white matter. To ensure the clinical usefulness of this algorithm with distributionally robust optimization, one must evaluate its performance and robustness against the criterion standard, ie, manual segmentation. Therefore, we compared automatic and manual segmentations of different brain structures on a series of super-resolution reconstruction fetal MR imaging volumes of fetal brain malformations.
MATERIALS AND METHODS
The education-support committee of the KU Leuven approved this study.
Data
This was a single-center retrospective study on fetal brain MR imaging, which was performed between October 1, 2016, and February 1, 2020, for CNS anomalies detected on prenatal ultrasound or for an increased risk of CNS anomalies (for diagnosis of included cases, see the Online Supplemental Data). All cases were selected from our data base on fetuses who were assessed because they were at increased risk for/suspected of having CNS abnormalities. Criteria for selection were severe brain malformations of different origins (eg, infectious, destructive, vascular, developmental, and so forth.) In addition, these cases could not have been included in the training data set. If multiple cases of the same pathology were present in the data base, the more severe one was selected. Images were acquired on a clinical 1.5T MR imaging system using a routine clinical protocol, without maternal sedation. This protocol includes T2-weighted single-shot turbo spin-echo sequences of the fetal brain in 3 orthogonal planes (see the Online Supplemental Data for MR imaging parameters), with repetition because of fetal motion when deemed necessary by the attending radiologist (M.A.). 2D slices were reconstructed to isotropic 3D volumes (resolution 0.8 × 0.8 × 0.8 mm) using the super-resolution reconstruction algorithm on a server in the hospital network.2
Method
Automatic Segmentation.
Isotropic 3D volumes of 16 fetuses were used as input of a CNN, which was trained using distributionally robust optimization for the fully automatic segmentations of the cerebellum, ventricular system, and white matter on both normal and abnormal brains.19 When we trained the algorithm, it was agreed that the segmentation of the ventricular system would include the lateral, third, and fourth ventricles with the cerebral aqueduct, cavum septum pellucidum, and cavum vergae, when present. The term “white matter” used throughout this article is for ease of use. What eventually becomes white matter consists, during fetal development, of multiple transient layers.20 Both the automatic and manual segmentations of what is referred to as white matter included these transient layers, more specifically the intermediate zone, cortical subplate, and ventricular zones.
There was no overlap between testing and training data in terms of subjects. The original training data set included 162 patients (124 controls without CNS abnormalities, 28 with spinal dysraphism, and 10 with other CNS abnormalities; see the Online Supplemental Data for more detailed information), with a gestational age range between 21 and 37 weeks.
Manual Segmentation.
The reference standard was set by manual segmentation of the selected brain structures, using the automatic segmentations as a starting point. Manual segmentations were performed using the software application ITK-SNAP (Version 3.8.0; www.itksnap.org).21 These structures were first segmented by a radiology resident (T.D.) and then reviewed and corrected by an experienced fetal radiologist (M.A).
Evaluation.
To compare the automatic and manual segmentations, we used quantitative methods and a descriptive discussion with reference to the underlying pathology. Quantitative comparison was performed with the Dice coefficient (DC) for volume overlap and the Hausdorff distance at the 95th percentile (Hausdorff 95%) between manual and automatic segmentation, because these were also the 2 metrics that were previously used in the original evaluation of the adopted CNN. We additionally calculated the volume difference between automatic and manual segmentations as an absolute value in Euclidean space. The volume difference was added because of the clinical relevance of volume measurements.
As is common in medical imaging analysis, for a surface-distance parameter, Hausdorff 95% was chosen (rather than at percentile 100).22 This makes more sense in our study because of the inevitable minor manual segmentation errors, as well as the few stray voxels that are rarely segmented by the CNN, which can even be located outside the skull. These result in a few extreme outliers that do not accurately represent the overall performance. We were particularly interested in measuring the statistical dispersion of the results as a way to evaluate the robustness of the algorithm, as well as to identify outlier values. To this end, we used the SD that is sensitive to outliers and the interquartile range (IQR) that is robust to outliers.22
Statistical analysis was performed using Python (https://www.python.org/) and Excel 2016 (Microsoft). To detect outliers, we defined these metric cutoff values as either lower than Q1 − (1.5 × IQR) or higher than Q3 + (1.5 × IQR), with Q1 and Q3 being the first and third quartiles. These outliers were descriptively discussed in the context of the underlying anomalies on a case-by-case basis.
RESULTS
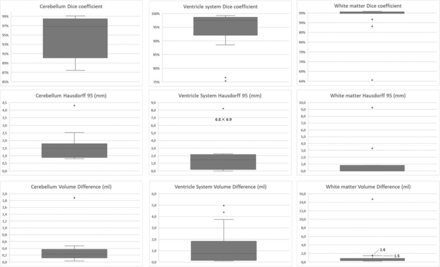
In 2 cases, the super-resolution reconstruction algorithm failed due to severe motion corruption; thus, these images were excluded. The gestational age in the remaining 14 cases was between 21.6 and 39.7 weeks (mean, 27.5 [SD, 4.4] weeks). For each of the 14 cases, the DC, Hausdorff 95%, and volume difference were calculated for the white matter, ventricle system, and cerebellum. Thus, 9 measurements were obtained per case, resulting in 126 measurements. The results of the comparison of automatic-to-manual segmentations are shown in Table 1; total volume and relative volume difference were added as a reference for the volume difference. The median DC for the white matter was 99.5%, which ranged between 64.5% and 99.8%. For the ventricular system, the median DC was 97.4% (range, 75.3%–99.3%). For the cerebellum, the median DC was 96.8% (range, 87.4%–99.1%). The median Hausdorff 95% of the white matter was 0.0 mm (range, 0.0–9.3 mm). For the ventricular system, the median Hausdorff 95% was 1.5 mm (range, 0.0–8.2 mm). For the cerebellum, the median Hausdorff 95% was 1.5 mm (range, 0.8–4.3 mm). The median volume difference for the white matter was 0.4 mL (range, 0.2–14.7 mL); for the ventricular system, it was 0.7 mL (range, 0.1–5.0 mL); and for the cerebellum, it was 0.2 mL (range, 0.0–1.9 mL). Note that the relative volume difference metrics based on all individual data (as in Table 1) is not necessarily the same as the relative comparison of the metrics of volume difference and total volume.
Evaluation of automatic segmentations for the 14 volumes of abnormal fetal brainsa
Of the above 126 measurements, there were 16 outliers in 5 fetuses as illustrated in the boxplots of the Figure.
Results of the automatic segmentations compared with the manual reference for the cerebellum, ventricular system, and white matter in our data set of fetuses with severe CNS anomalies. The metrics used are the DC, Hausdorff 95, and volume difference. Outliers are represented by dots in the boxplots, defined as Q1 – (1,5 × IQR) or Q3 + (1,5 × IQR); Q1 and Q3 are the first and third quartiles and IQR is the interquartile range. Note that different scales are used for optimal visualization.
DISCUSSION
In this data set of fetuses scanned for CNS abnormalities, we found an overall excellent correlation of automatic and manual segmentations of the white matter, ventricular system, and the cerebellum. This was supported by the high DC, low Hausdorff 95%, and small volume difference. On the basis of our own evaluation of the algorithm, using our data set with abnormal fetal brains, we additionally compared the performance of the CNN with results reported earlier using other methods and data sets (Table 2). DC values of >70% are usually considered consistent with a satisfactory level of agreement between 2 segmentations.23 This comparison obviously provides only an indication because those studies were performed on other data sets with either healthy fetuses and/or fetuses with different pathologies.
Automatic segmentation performance on our testing data set (trained using distributionally robust optimization) compared with other methods trained and evaluated with different data setsa
Of the 126 measurements, there were 13% outliers (n = 16); and artifacts such as the partial volume effect could account for only a minor contributing factor. Conversely, most of these outliers were present in fetuses with very specific anatomic changes. Thus, the outliers are further descriptively discussed on the basis of the underlying condition, hereby identifying when the algorithm makes segmentation errors. This feature emphasizes the limitation of our study, because the number of cases of each pathology is rather small (Online Supplemental Data). As a hypothesis for further research to improve the algorithm, one would need to train the algorithm with additional images with fetal pathologies similar to those in the erroneous cases and at a similar gestational age, to rule out the effect of brain development. This hypothesis is possibly strengthened by the finding that none of the automatic segmentations of the 2 Chiari II cases had outlier values, possibly due to the large portion of spinal dysraphism cases in the training data set (28 of the 162 cases).
When we looked at the outlier values of our metrics, we found the most extreme outliers for the white matter and ventricular system in a case of an intracranial hemorrhage, which happened to have a signal intensity similar to that of the developing white matter at the time of imaging. The hemorrhage followed the convexity of the skull bilaterally, deviating the parenchyma medially, thus occupying the space where the white matter is typically found (Online Supplemental Data). This change caused errors in the automatic segmentations, which was to be expected because there were no cases with hemorrhages in the training data set. Because we had a postmortem scan available at the time of manual segmentation, we could verify our manual segmentation and determine that the automatic segmentation algorithm included parts of the hemorrhage in the segmentation of the white matter (see Online Supplemental Data for MR imaging parameters). In the same case, the CNN erroneously included parts of the hemorrhage, extra-axial CSF, as well as porencephalic changes in the segmentation of the ventricular system (Online Supplemental Data). These oversegmentations correspond to the outliers of the DC, Hausdorff 95%, and volume difference for both white matter and the ventricular system.
There was only 1 case with outliers in the segmentation of the cerebellum. This can be explained by the inherent changes due to the pathology present in this case, ie, an aqueductal stenosis. Due to the stenosis, there is a dilation of the supratentorial ventricular system with accompanying mass effect, which, in turn, alters the configuration of the posterior fossa. More precisely, this scenario leads to a redistribution of the pericerebellar CSF and a reduced space between the cerebellum and the tentorium. Presumably, this altered configuration causes undersegmentation of the anterior lobe by the CNN (Online Supplemental Data). The cerebellar folia in the region of the vermis were also undersegmented, due to the partial volume effect brought on by the different distribution of the surrounding CSF. This feature translated into outlier values in Hausdorff 95% and volume difference.
In the same fetus, there were also outliers for the white matter segmentation (both for DC and Hausdorff 95%). The aqueductal stenosis caused dilation of the supratentorial ventricular system and secondary white matter thinning. This result created a thin and irregular segmentation that caused variable DCs, because the DC is inherently highly susceptible to these irregularities (Online Supplemental Data). In addition, this fetus was an outlier due to its advanced gestational age (39.7 weeks), at which time there is advanced gyrification and a decrease in the subcortical plate tissue throughout the brain, with physiologic remnants in different areas. The combination of both physiologic processes makes the white matter more heterogeneous and probably more difficult for the CNN trained algorithm to segment correctly. The algorithm we used was trained on younger fetuses (21–37 weeks) because these are more commonly scanned; therefore, it had no prior experience with fetuses of that advanced age.
There were also outliers for the ventricular system segmentation in 2 of the 3 cases with (partial) corpus callosum agenesis, in which the CNN makes an oversegmentation of the extraventricular CSF in the interhemispheric cistern (Online Supplemental Data). This finding is likely due to the algorithm falsely assuming the presence of a cavum septum pellucidum. This structure is present in healthy fetuses, but absent in corpus callosum agenesis. As mentioned earlier, it was agreed in advance to include the cavum septum pellucidum in the segmentation of the ventricular system (both when training the CNN, as well as for the manual segmentation comparison). This inclusion was because the cavum septum pellucidum is a normal structure in normal fetal brains, though we acknowledge it is not part of the ventricular system. Furthermore, in the training data set, we included several cases with spinal dysraphism in which the cavum septum pellucidum can be absent or incomplete.24,25 In corpus callosum agenesis, the lateral ventricles are generally more widely spaced and the third ventricle is dilated and may communicate with the interhemispheric fissure. Therefore, we hypothesize that the algorithm recognizes the fluid-filled space or associated cyst between the hemispheres as the cavum septum pellucidum. In one of these 2 cases, a borderline outlier was also seen in the white matter volume difference, presumably because of the partial volume effect in the narrow parts of the lateral ventricles, in turn causing lower-signal-intensity voxels of the CSF to be wrongfully included (Online Supplemental Data).
Finally, there was an outlier in the DC of the white matter in a case with idiopathic dilation of the lateral ventricles. We attribute this to the previously mentioned factors of thin and irregular white matter due to ventriculomegaly (Online Supplemental Data). We have focused on the outliers because they are the most interesting for further development of segmentation algorithms. For completion, we have also added an example of a case in which the algorithm performs well; thus, there is a good correlation of the automatic and the manual segmentations (Online Supplemental Data).
Note that the commissures were not added to the white matter segmentation, even though we acknowledge that commissural fibers are white matter. This decision was due to additional brain structures being added in later versions of the algorithm, which include commissural fibers such as the corpus callosum.
Another potential limitation is that the manual segmentation was performed using the automatic segmentation as a starting point. This step was to simultaneously identify potential errors in the automatic segmentation, being important to the engineers involved in the development and further optimization of the algorithm. To minimize the potential effect on the statistical results, we reviewed the manual segmentations and an experienced fetal radiologist (M.A.) corrected them after the initial manual segmentation by a radiology resident (T.D.). As a final limitation, we acknowledge the small number of cases of each pathology, which has been mentioned before in this section.
CONCLUSIONS
We demonstrated an overall excellent correlation between the automatic segmentation by the CNN and the ground truth manual segmentations in a new clinical data set, consisting exclusively of cases with a variety of severe brain abnormalities. Additionally, our results suggest the need to include enough cases with a diverse spectrum of pathologies and a broad age range when training these algorithms to prevent errors. The remaining errors emphasize the need to perform a manual check and look for occasional faults of the algorithm due to severe or rare abnormalities or induced by artifacts. Nevertheless, the vast time savings would suggest that this algorithm is very useful for managing clinical data sets with a variety of pathologies.
Footnotes
Disclosure forms provided by the authors are available with the full text and PDF of this article at www.ajnr.org.
References
- Received February 6, 2022.
- Accepted after revision February 6, 2023.
- © 2023 by American Journal of Neuroradiology