Abstract
BACKGROUND AND PURPOSE: Communication gaps exist between radiologists and referring physicians in conveying diagnostic certainty. We aimed to explore deep learning–based bidirectional contextual language models for automatically assessing diagnostic certainty expressed in the radiology reports to facilitate the precision of communication.
MATERIALS AND METHODS: We randomly sampled 594 head MR imaging reports from an academic medical center. We asked 3 board-certified radiologists to read sentences from the Impression section and assign each sentence 1 of the 4 certainty categories: “Non-Definitive,” “Definitive-Mild,” “Definitive-Strong,” “Other.” Using the annotated 2352 sentences, we developed and validated a natural language-processing system based on the start-of-the-art bidirectional encoder representations from transformers (BERT), which can capture contextual uncertainty semantics beyond the lexicon level. Finally, we evaluated 3 BERT variant models and reported standard metrics including sensitivity, specificity, and area under the curve.
RESULTS: A κ score of 0.74 was achieved for interannotator agreement on uncertainty interpretations among 3 radiologists. For the 3 BERT variant models, the biomedical variant (BioBERT) achieved the best macro-average area under the curve of 0.931 (compared with 0.928 for the BERT-base and 0.925 for the clinical variant [ClinicalBERT]) on the validation data. All 3 models yielded high macro-average specificity (93.13%–93.65%), while the BERT-base obtained the highest macro-average sensitivity of 79.46% (compared with 79.08% for BioBERT and 78.52% for ClinicalBERT). The BioBERT model showed great generalizability on the heldout test data with a macro-average sensitivity of 77.29%, specificity of 92.89%, and area under the curve of 0.93.
CONCLUSIONS: A deep transfer learning model can be developed to reliably assess the level of uncertainty communicated in a radiology report.
ABBREVIATIONS:
- AUC
- area under the receiver operating characteristic curve
- BERT
- bidirectional encoder representations from transformers
- BioBERT
- biomedical variant of BERT
- ClinicalBERT
- clinical variant of BERT
- NLP
- natural language processing
- QC-RAD
- qualifying certainty in radiology reports
The American College of Radiology has stressed a critical need for “precision communication” in radiologic reports,1,2 and clarity has also been recognized by referring physicians as a key quality metric of radiologic reports.3 However, there are communication gaps when the referring physician may interpret the radiologist’s textual expressions that convey diagnostic uncertainty as different from what was intended.4⇓-6
A standardized lexicon for diagnostic certainty7 has been proposed to address this challenge. However, using a restricted lexicon can only help with lexion-level “one word at a time” interpretation, while diagnostic uncertainty in the radiology report is typically context-dependent. It means that the same term may have different interpretations based on how differential diagnoses are reported in the context. For instance, “likely to be Arnold Chiari I malformation” indicates a mildly certain diagnosis, while “likely differential considerations include demyelinating/inflammatory processes” indicates uncertainty in the diagnosis. Additionally, standardized lexicons will not mitigate the problem of overusing “hedge” words when the diagnosis could be more certain.8
Natural language processing (NLP), an artificial intelligence technology that analyzes free texts to understand underlying semantics, has been widely applied to radiology research for automatic identification and extraction of clinically important information.9 However, little work has been done on applying NLP to assess the diagnostic certainty in radiology reports beyond the lexicon level (eg, hedging cue terms). NegBio (https://github.com/ncbi-nlp/NegBio)10 was developed to detect negation and uncertainty in radiology reports, but the rule-based system highly depends on identifying relevant hedging terms only, without considering linguistic contexts. It only performs binary identification regarding whether a finding is uncertain or not. Other research has attempted to analyze the certainty of scientific statements in the biomedical literature through conventional machine learning methods (eg, conditional random fields,11 support-vector machines12⇓-14), but the ground truth annotation used to train these systems is based on which hedging expressions appear in the sentence, and these hedging expressions do not provide a reliable metric in diagnostic radiology reporting.
Recent innovations in deep learning technology provide improved NLP performance in radiology-related research.15 Exploring state-of-the-art deep learning approaches to develop a reliable NLP system to qualify the context-aware certainty in radiology reporting has great potential to facilitate communications between radiologists and referring physicians. The goal of this study is to investigate the potential of deep learning NLP techniques for qualifying the certainty expressed in the Impression sections of radiology reports (QC-RAD). Specifically, our goal is the following:
Establish an NLP system for capturing contextualized certainty semantics at the sentence level in radiology reports and build the first annotated data of its kind
Develop a deep transfer learning approach so that knowledge representation learned from a very large universal textual data set can be transferred through fine-tuning the pretrained neural networks
Conduct experiments with 3 variants of bidirectional encoder representations from transformers (BERT) models with demonstrated promising results.
MATERIALS AND METHODS
Figure 1 shows the overview of the QC-RAD system, and we will describe each component in the following subsections.
Overview of the QC-RAD system workflow.
Data Annotation
In this institutional review board–exempt quality improvement project, we initially randomly selected 1500 brain MRIs performed at a single academic medical center (UMass Memorial Medical Center). During the data-cleaning process, we transformed the reports into plain texts and extracted the Impression sections, which contain free-form texts written by the radiologists. We then loaded them into the extensible Human Oracle Suite of Tools (eHOST; https://github.com/chrisleng/ehost)16 for context-aware certainty annotation. The eHost is versatile for annotation tasks and has been used by several institutions and projects for a variety of tasks, including i2b2: Informatics for Integrating Biology & the Bedside17 and the Consortium for Health Care Informatics Research18 projects.
We asked 3 board-certified radiologists (S.J.B., M.J.M., and N.A.) to read sentences from the Impression sections and to assign each sentence 1 of the 4 certainty categories shown in Table 1. We define “diagnostic findings” as a diagnostic opinion regarding a specific disease or other condition.19 The certainty category is not only dependent on the hedging terms used but is also based on the holistic context expressed in the sentence. In contrast to previous studies, we did not try to distinguish different hedging terms or phrases from their different levels of certainty because they are perceived so differently by physicians, radiologists, and patients.5,6 In our annotation guideline, we compiled a list of hedging terms (Online Supplemental Data), and using any of them is considered one of the factors contributing to uncertainties (defined in Table 1).
Diagnostic certainty of diagnosis in the Impression section of a radiology report—categories for annotation
Initially, the 3 annotators each reviewed 30 head MR imaging reports and then went through an iterative process of adjusting the guideline and amending the annotations to reach consensus. Each annotator then independently annotated an additional 24 MR imaging reports according to the finalized annotation guideline (derived from the consensus review of the initial 30 reports). The interrater agreement was calculated (0.74) by mean pair-wise Cohen κ statistics, which showed substantial strength of agreement across annotators.20 Finally, each annotator annotated 180 reports, resulting in a total of 594 reports.
The annotated data were then analyzed for certainty qualification, as follows: We performed word tokenization and sentence boundary identification on all the sentences from the Impression section of the radiology reports. We then removed sentences that contained fewer than 4 words (76 sentences) because these short sentences are typically noise caused by sentence-splitting errors, resulting in 2352 sentences in total for further analysis; 88.7% of the sentences were <25 words in length, with a mean of 14 (SD, 9.5). We then split the annotated certainty data into training data (80%), validation data (10%), and testing data (10%). The training and validation data were used for fine-tuning, and the test data were used as heldout (unseen) data to evaluate the final performance of the system. The data statistics on 3 datasets are shown in Table 2.
Data statistics of the 3 data setsa
Deep Transfer Learning
We formulated the certainty assessment task into a multiclass sentence-classification problem and exploited NLP techniques to capture fine-grained semantics for classifying each sentence into 1 of the 4 categories defined in Table 1. Recent progress in NLP has been driven by using deep learning approaches,21 and different deep learning architectures have been applied for text classification, which typically can be grouped into 2 model families: Convolutional neural networks are good at extracting local and position-invariant pattern features,22 while recurrent neural networks are shown to perform better in modeling long dependencies among texts.23 All those approaches require large amounts of labeled data to reliably estimate the numerous model parameters; however, compared with general domains, annotated data are more difficult and expensive to obtain in clinical domains because they require subject matter expertise for high-quality annotation.
Deep transfer learning24 makes it possible to harness the power of deep neural architecture when only limited labeled data are available. In this study, we explored the state-of-the-art NLP transferring learning model BERT,25 which was developed by Google Artificial Intelligence and demonstrated breakthrough performance improvement in a variety of NLP tasks. BERT is a contextualized word-representation model based on a masked language model and pretrained using bidirectional transformers,26 and it can take into account sequential dependencies among words in a sentence for a semantically meaningful representation. More information about the BERT can be found in the Online Supplemental Data.
Preprocessing.
We extracted data from the annotation tool and performed sentence segmentation using the Natural Language Toolkit (https://www.nltk.org/)27 so that each sentence was paired with a certainty label described in Table 2. We then used the WordPiece tokenizer (https://www.paperswithcode.com/method/wordpiece)28 for tokenization. It breaks each word down into its prefix, root, and suffix (subwords) in order to mitigate the out-of-vocabulary issue. For instance, “infarction” will be tokenized as 3 subwords: “in,” “##far,” and “##ction.” To be compatible with BERT input format, we added the “[CLS]” token at the beginning of each sentence, and the “[SEP]” token at the end of each sentence.
Model Training (Fine-tuning).
To address our sentence-classification problem, we added a drop-out regularization29 and a softmax classifier layer on top of the pretrained BERT layer. In our work, we adopted 3 variants of pretrained BERT models: 1) The BERT-base model (https://github.com/google-research/bert) consists of an encoder with n (n = 12 in Fig 2) layers of transformer blocks and was pretrained using BookCorpus (https://github.com/soskek/bookcorpus) and Wikipedia;25 2) BioBERT (https://github.com/dmis-lab/biobert)30 was initialized using the BERT-base and was pretrained using BookCorpus, Wikipedia, PubMed abstracts, and PubMed Central full text articles; 3) ClinicalBERT (https://github.com/EmilyAlsentzer/clinicalBERT)31 was initialized using BioBERT and pretrained using around 2 million clinical notes in the MIMIC-III database (https://physionet.org/content/mimiciii/1.4/).32 All 3 model variants shared the same architecture with the BERT-base model, which consists of 12 layers of transformer blocks with a hidden size of 768, and 12 self-attention heads. We fine-tuned all the layers in the model-training process. The 3 model variants shared the following hyperparameters: drop-out probability of 0.1 and maximum sequence (subwords) length of 80. The optimal batch size, learning rate, and number of epochs were chosen using the validation data (see the Results section).
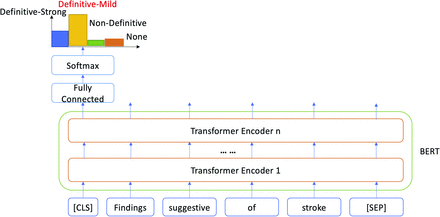
Illustration of using BERT for certainty classification. The input “Findings suggestive of stroke” was classified as “Definitive-Mild.”
Model Evaluation.
We evaluated the performance QC-RAD using standard metrics: Sensitivity, specificity, and area under the receiver operating characteristic curve (AUC). For aggregated evaluation across 4 certainty categories, we use the macro-average value, which calculates each metric (sensitivity, specificity, or AUC) independently for each category and then takes the average. We prefer macro-average to micro-average because we value the ability of QC-RAD to perform equally well across different categories. In the context of imbalanced data in which the majority category has many more samples than other categories, micro-average will be biased toward the dominating majority categories, while macro-average is less sensitive and considers each category equally.
Figure 2 shows an example. The input sentence “Findings suggestive of stroke” is composed of a sequence of 4 words. [CLS] and [SEP] are added prior to being fed into the BERT model. We first initialize n transformer encoders (n = 12 orange blocks in Fig 2) using the pretrained BERT model, and all the parameters, including the fully connected layer, will be fined-tuned through supervised learning using the labeled data (see the “Data Annotation” section) for certainty classification. Through a multilayer deep neural network architecture (encoder) in BERT, each input token will be transformed to a final output embedding (vector representation). For this sentence-classification task, we use only the final hidden state of the first token [CLS], which is considered aggregated sentence representations, and feed it into a fully connected layer to obtain a probability distribution across 4 certainty categories through the softmax function.
RESULTS
We reported the classification performance against the reference standard categories assigned by radiologists. The aggregated results are presented with the macro-average across 4 categories defined in Table 1. For 3 pretrained language models, we used grid-search to optimize the batch size (range in 24, 32, and 64) and learning rate (range in 0.000005, 0.00001, 0.00003, and 0.00005) during the fine-tuning process for a fair comparison. The number of epochs for fine-tuning training was selected on the basis of the peak AUC score on the validation data.
Comparative Performance among 3 BERT Models (Validation Data)
We compared the performance on the certainty classification task using the 3 pretrained BERT models, and the results are shown in Table 3. The BioBERT model obtained the best macro-AUC of 0.931, and the BERT-base yielded the best macro-sensitivity of 79.46% and specificity of 93.65%, while ClinicalBERT achieved the relatively lower macro-sensitivity of 78.52% compared with the other 2 models (Table 3).
Performance comparison among 3 BERT variants (with their optimal parameters) on the validation data set
Performance Curve in the Fine-tuning Process (Validation Data)
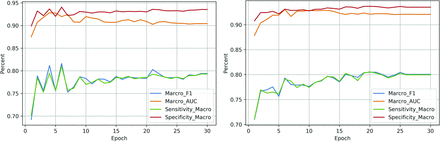
Figure 3 shows the performance curve of the BERT-base (left) and BioBERT (right) across the number of epochs during the fine-tuning process. Here, we also show the F1 score, which is the harmonic mean of the positive predictive value and sensitivity. We observed similar trends on both models. With fine-tuning, all the performance metrics increased initially and plateaued after approximately 5 epoch trainings.
Performance curve across the number of fine-tuning epochs. The left figure is for BERT, and the right one is for BioBERT.
Performance on the Test Data
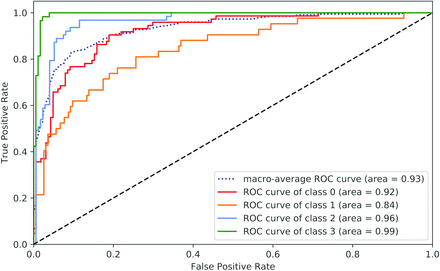
On the basis of the evaluation results (AUC scores) on the validation data, we chose the best system (BioBERT) and applied it to the test data as shown in Table 4. The system performs the best on the “Other” category, with the highest sensitivity of 98.31%, specificity of 97.19%, and AUC of 0.994. Among the other 3 categories, the system obtained the highest sensitivity for Non-Definitive (76.71%), the highest specificity for Definitive-Strong (95.4%), and the highest AUC for Definitive-Strong (0.964). Overall, it obtained the macro-average sensitivity of 77.29%, specificity of 92.89%, and AUC of 0.93 on the heldout unseen data. Although the Non-Definitive class has a lower AUC score than Definitive-Strong, the sensitivity of Non-Definitive was better than that of Definitive-Strong (76.71% versus 74.6%) as shown in Table 4. In Fig 4, receiver operating characteristic curves of Definitive-Strong (class 2) and Other (class 3) are closer to the ideal spot (the closer to the upper left corner, the better).
System performance of BioBERT on the test data seta
Receiver operating characteristic curves of individual classes on the test data set. Class 0 = Non-Definitive, class 1 = Definitive-Mild, class 2 = Definitive-Strong, and class 3 = Other.
Error Analysis
We conducted error analysis on the validation data, and the confusion matrix is shown in Table 5. Rows represent the truth label assigned by domain experts, and columns indicates the system predictions. It shows that only 1 (1.7%) sentence in the Other category was wrongly classified as Non-Definitive, which explains the high performance of this category in Table 4. On the basis of the definition of the “Other” category, it covers a narrow scope of semantics, and it is easy for the system to pick up the representative patterns (eg, “follow-up” is a reliable indicator for recommendations). The top 3 error patterns are the following: 1) Non-Definitive → Definitive-Mild (11 of 73, 15%), 2) Definitive-Mild → Non-Definitive (11 of 41, 26.8%), and 3) Definitive-Strong → Non-Definitive (11 of 63, 17.5%). Those error patterns suggest that the Non-Definitive category is more challenging to distinguish from the other 2 definitive categories.
Confusion matrix among different categories
DISCUSSION
We demonstrated that the interrater agreement of subject matter experts on certainty interpretations when considering both hedging term usage and surrounding linguistic contexts is excellent. Building on this ground truth, we then demonstrated that deep transfer learning shows great potential for unlocking contextualized semantics for certainty assessment of radiology reports using limited annotated data. Our novel QC-RAD system holds the potential to facilitate precision communication of imaging findings, as well as to serve as a new quality measure in radiology reporting.
Multiple publications have stressed the importance of the accurate conveying of diagnostic certainty in the radiology report. Most recently, a study developed a certainty scale, specifying recommended and nonrecommended certainty terms. The adoption of such a scale significantly increased the proportion of recommended certainty terms during a voluntary period.33 Similar to the idea of a standardized certainty lexicon,7 this approach is limited to term-level certainty, without taking into account contextual semantics. In contrast to prior work that has used NLP to simply identify the instances of these predefined terms and to count the frequency of specific, predefined hedging terms, our work has developed a deep learning–based NLP algorithm that will read and interpret the level of certainty conveyed in the free text, including surrounding contexts.
Our study categorized the certainty of each sentence in the Impression section of radiology reports into different certainty categories. We showed that our automated categorization scheme has strong operating characteristics compared with a ground truth based on the radiologists’ consensus. The sentence-level certainty categorization is a first step toward a more general quantification of diagnostic uncertainty conveyed in radiology reports.
Deep learning approaches have shown breakthrough results in many tasks, but it is challenging to train a reliable deep neural network with limited annotation data in a specific domain. Pretrained language models, such as BERT, greatly alleviate this problem by training a deeply bidirectional language representation in an unsupervised manner using only a plain text corpus. By leveraging this universally learned knowledge, we performed the fine-tuning using the task-specific annotated data so that the learned deep encoder can be adapted to better fit our target task. Our experiments show promising results on all 3 variants of the BERT models, leading to the best macro-average AUC score of 0.93 on the unseen test data.
Among the 3 variant models of BERT, though ClinicalBERT was pretrained using clinical notes, it did not show any advantage compared with the other 2 models (BERT and BioBERT) as it did in other benchmark tasks.31 This finding is possibly because clinical notes often contain ill-formed, nongrammatic sentences, arbitrary abbreviations, and typographic errors, which are less likely to be present in the Impression section of radiology report. Therefore, the expanded knowledge from clinical notes may not necessarily benefit the current task and could potentially introduce noise. BioBERT did show a slight overall performance gain in terms of the AUC score; however, at a certain threshold, BERT-base outperformed it on the basis of sensitivity and specificity.
The Impression of the radiology report reflects the radiologist’s interpretation of actionable findings on the imaging study. Sometimes the diagnoses are inherently uncertain by imaging alone. However, when there is near-certainty about the diagnosis, radiologists should convey confidence.8 Our ultimate goal is to provide the radiologist with a real-time, automatic measurement of the level of diagnostic certainty in their report before signing. Thus, the radiologists will have objective information about the level of certainty that they are conveying. The current system classifies 1 sentence into a discrete certainty category. In future work, we will expand the system by combining the discrete certainty category with the probability that the certainty level is correct. Once this expanded system has been developed and appropriately validated, the quantification of the certainty level conveyed in the radiology report may be used as a quality metric to evaluate radiologists’ performance.
There are limitations in this study. First, the data size used for this study is relatively small, yet the high-quality annotation and transfer-learning strategy enable the system to learn efficiently, yielding promising results on the withheld testing data. Leveraging a larger amount of unlabeled data (eg, unsupervised representation learning34) would potentially further improve the performance of the system, which we will explore for future work. Second, we developed and evaluated the QC-RAD system using head MR imaging radiology reports only, and its generalizability to other radiologic specialties needs further investigation. However, because our system is fully trainable and does not depend on any heuristic rules, we speculate that it can be easily generalized to other radiology subspecialties through active-learning35 and domain-adaptation learning36 techniques. Third, cross-institutional external validation on the performance of the system and ground truth consensus is needed to verify the overall generalizability of the study, which we will pursue in the near future. We did conduct an external validation within our institution. Specifically, we randomly sampled a new set of 40 MR imaging head reports from 4 new neuroradiologists (10 reports each) who were not covered by our original data collection. We asked the same 3 radiologists to assign 1 of the 4 certainty categories to all the 132 sentences from the Impression sections of these 40 reports. We found that the interannotator agreement remained very high with a mean pair-wise κ score of 0.761. We chose the annotations from the annotator who agreed most with the other 2 as ground truth and evaluated the performance of QC-RAD on this new data set, achieving the macro-sensitivity of 84.01%, macro-specificity of 93.59%, and a macro-AUC of 0.945. This validation experiment demonstrates great generalizability of our QC-RAD system and ground truth consensus.
CONCLUSIONS
We developed and validated a deep transfer learning system, QC-RAD, to automatically assess the level of certainty in head MR imaging reports. Experimental results demonstrated that QC-RAD can effectively unlock contextualized semantics of free-text reporting language for assessment of diagnostic certainty in radiology reports, holding the potential to facilitate precision communication of imaging findings between radiologists and referring physicians.
ACKNOWLEDGMENTS
The authors thank Dr Alexander A. Bankier for the insightful discussions and comments on the manuscript.
Footnotes
This work was supported by a research fund from the Department of Radiology, UMass Medical School.
Disclosures: Feifan Liu—RELATED: Grant: radiology department research funding; UNRELATED: Patents (Planned, Pending or Issued): A provisional patent was filed; Grants/Grants Pending: National Institute of Mental Health, National Library of Medicine, National Science Foundation, National Cancer Institute.* Steven J. Baccei—UNRELATED: Royalties: UpToDate, Comments: author of an UpToDate article; royalties paid biannually; Payment for Development of Educational Presentations: Conserus Healthcare, Comments: patient safety educational program. Catarina I. Kiefe—UNRELATED: Grants/Grants Pending: National Institute of Mental Health, National Heart, Lung and Blood Institute, National Center for Advancing Translational Sciences, National Cancer Institute.* *Money paid to the institution.
References
- Received November 25, 2020.
- Accepted after revision May 19, 2021.
- © 2021 by American Journal of Neuroradiology