Abstract
BACKGROUND AND PURPOSE: WHO grade II gliomas are divided into three classes: isocitrate dehydrogenase (IDH)-wildtype, IDH-mutant and no 1p/19q codeletion, and IDH-mutant and 1p/19q-codeleted. Different molecular subtypes have been reported to have prognostic differences and different chemosensitivity. Our aim was to evaluate the predictive value of imaging phenotypes assessed with the Visually AcceSAble Rembrandt Images lexicon for molecular classification of lower grade gliomas.
MATERIALS AND METHODS: MR imaging scans of 175 patients with lower grade gliomas with known IDH1 mutation and 1p/19q-codeletion status were included (78 grade II and 97 grade III) in the discovery set. MR imaging features were reviewed by using Visually AcceSAble Rembrandt Images (VASARI); their associations with molecular markers were assessed. The predictive power of imaging features for IDH1-wild type tumors was evaluated using the Least Absolute Shrinkage and Selection Operator. We tested the model in a validation set (40 subjects).
RESULTS: Various imaging features were significantly different according to IDH1 mutation. Nonlobar location, larger proportion of enhancing tumors, multifocal/multicentric distribution, and poor definition of nonenhancing margins were independent predictors of an IDH1 wild type according to the Least Absolute Shrinkage and Selection Operator. The areas under the curve for the prediction model were 0.859 and 0.778 in the discovery and validation sets, respectively. The IDH1-mutant, 1p/19q-codeleted group frequently had mixed/restricted diffusion characteristics and showed more pial invasion compared with the IDH1-mutant, no codeletion group.
CONCLUSIONS: Preoperative MR imaging phenotypes are different according to the molecular markers of lower grade gliomas, and they may be helpful in predicting the IDH1-mutation status.
ABBREVIATIONS:
- AUC
- area under the receiver operating curve
- IDH1
- isocitrate dehydrogenase 1
- LASSO
- Least Absolute Shrinkage and Selection Operator
- LGG
- lower grade glioma
- VASARI
- Visually AcceSAble Rembrandt Images
- WHO
- World Health Organization
Recently, the 2016 World Health Organization Classification of Tumors of the Central Nervous System was published. In the new classification, both genotype and phenotype are combined to define each diagnostic category of diffuse gliomas.1 For the diagnosis of lower grade gliomas (LGGs), which are World Health Organization (WHO) grade II and III gliomas, the isocitrate dehydrogenase (IDH) mutation status, and 1p/19q-codeletion status are combined with the histologic phenotype, and the genotype takes precedence over the histologic phenotype in cases of discordance. Recent studies of genomic analysis found that diffuse gliomas may have distinct clinical behavior according to their molecular marker status.2,3 The results of these studies have been reflected in the 2016 World Health Organization Classification of Tumors of the Central Nervous System, which is based on the integrated diagnosis, combining phenotypic and genotypic classifications.4
The molecular subtypes of diffuse gliomas are divided into 3 classes: IDH wild type, IDH mutant and no 1p/19q codeletion, and IDH mutant and 1p/19 codeleted.1 Different molecular subtypes have been reported to have prognostic differences and different chemosensitivity.3,5 Thus, predicting the molecular subtype of LGGs preoperatively by MR imaging may aid in predicting the prognosis and planning the treatment strategy.
The recently developed lexicon, Visually AcceSAble Rembrandt Images (VASARI), is a tool for describing the baseline imaging features of human gliomas with MR imaging.6 Using specific guidelines and controlled lexicon, the feature set is designed to comprehensively describe the tumor in consistent and reproducible terms. The imaging features recorded have been reported to be clinically meaningful, widely available, reproducible, and biologically relevant.6⇓–8
The purpose of this study was to evaluate the predictive value of imaging phenotypes assessed with the VASARI lexicon for the molecular classification of LGGs preoperatively.
Materials and Methods
Patient Population
The Severance Hospital institutional review board waived patient consent for this retrospective study. Between January 2007 and February 2017, three hundred sixty-six patients with pathologically diagnosed LGGs were included in this study. Inclusion criteria were as follows: 1) WHO grade II and III gliomas confirmed by histopathology; and 2) patients who underwent preoperative MR imaging. Exclusion criteria were the following: 1) an unknown IDH1-mutation status; 2) patients with a previous history of brain tumors; 3) patients with incomplete MR imaging sequences or suboptimal image quality for the VASARI lexicon review; and 4) patients younger than 18 years of age. The extent of tumor resection was classified as total, subtotal (<100% and ≥75% of gross total removal), or partial (<75% of gross tumor removal) resection or stereotactic biopsy only on the basis of the surgeon's intraoperative impressions in conjunction with postoperative MR imaging findings. The study population was chronologically divided into 2 sets. First, 175 consecutive patients diagnosed as having LGGs between January 2007 and May 2016 were included in the discovery set, and 40 consecutive patients diagnosed as having LGGs between June 2016 and February 2017 were included in the independent validation set. The flow chart of the study population is shown in On-line Fig 1. Patient characteristics of the discovery and validation sets are shown in On-line Table 1. The mean interval between the MR imaging examination and the operation was 10.73 ± 12.49 days.
Peptide nucleic acid–mediated clamping polymerase chain reaction and immunohistochemical analysis were performed to detect an IDH1-R132H mutation.9 In the immunohistochemical analysis, the monoclonal antibody H09 was used, and the degree of IDH1-R132H staining was determined as positive in patients with any stained cells.10 If immunohistochemical staining results were negative, we confirmed the IDH1 status by peptide nucleic acid–mediated clamping polymerase chain reaction. Fluorescent in situ hybridization analysis was used to investigate the 1p/19q codeletion.11
MR Imaging Protocol
Preoperative MR imaging was performed with a 3T MR imaging scanner (Achieva; Philips Medical Systems, Best, Netherlands) with an 8-channel sensitivity-encoding head coil. The preoperative MR imaging protocol included T1-weighted (TR/TE, 1800–2000/10–15 ms; FOV, 240 mm; section thickness, 5 mm; matrix, 256 × 256), T2-weighted (TR/TE, 2800–3000/80–100 ms; FOV, 240 mm; section thickness, 5 mm; matrix, 256 × 256), and fluid-attenuated inversion recovery images (TR/TE, 9000–10,000/110–125 ms; FOV, 240 mm; section thickness, 5 mm; matrix, 256 × 256). 3D contrast-enhanced T1-weighted images (TR/TE, 6.3–8.3/3.1–4 ms; FOV, 240 mm; section thickness, 1 mm; and matrix, 192 × 192) were acquired after administering 0.1 mL/kg of gadolinium-based contrast material (gadobutrol, Gadovist; Bayer Schering Pharma, Berlin, Germany). Diffusion tensor imaging was performed with b-values of 600 and 0 s/mm2, 32 directions, and the following parameters: TR/TE, 8413.4/77 ms; FOV, 220 mm; section thickness, 2 mm; and matrix, 112 × 112.
Imaging Analysis
Two neuroradiologists (S.S.A. and Y.W.P. with 10 and 5 years of experience, respectively), blinded to the molecular data, independently reviewed the MR imaging scans for tumor size, location, and tumor morphology using a standardized imaging feature set, VASARI. Discrepancies between the 2 radiologists were settled by consensus. The VASARI lexicon for MR imaging annotation consists of 26 imaging descriptors based on different MR imaging features. The exact description of all the features can be found at the Cancer Imaging Archive of the National Cancer Institute (https://wiki.cancerimagingarchive.net/display/Public/VASARI+Research+Project), which includes imaging features related to lesion location, morphology of the lesion substance, morphology of the lesion margin, alterations near the lesion, and remote alterations.
Statistical Analysis
The interrater agreement for the imaging features was assessed by using the Cohen κ coefficient test. The Student t and Pearson χ2 tests were performed to evaluate the association between the imaging features and IDH1-mutation status in the discovery group, including the WHO grade II and III subgroups. Next, because the number of significant imaging features was relatively large compared with the number of patients when comparing the IDH1-wild type and IDH1-mutant groups, we used the regularization method to assess the predictive power of the imaging features based on the Least Absolute Shrinkage and Selection Operator (LASSO), which reduces the potential risk of overfitting or false discovery. We used 10-fold cross-validation to find the optimal regularization parameter for LASSO. We estimated the area under the receiver operating characteristic curve (AUC) to assess the predictive ability of variables by selecting significant variables based on LASSO. The 10-fold cross-validated AUC is the average of the predictive AUC of 10 validation datasets generated by the cross-validation process. Then, using the significant variables from the discovery set, we obtained the AUC in the validation set. The 5-fold cross-validated AUC was estimated to assess the predictive ability of variables by LASSO in the grade II and III subgroups.
The Student t and Pearson χ2 tests were performed to evaluate the association between the imaging features and 1p/19q-codeletion status in the IDH1-mutant subgroup of the discovery group. LASSO was not performed to predict the 1p/19q-codeletion status in the IDH1-mutant subgroup because there were few significant parameters according to the 1p/19q-codeletion status. Statistical analysis was performed by with R statistical and computing software (http://www.r-project.org). Statistical significance was set at P < .05.
Results
Characteristics of the 175 enrolled patients in the discovery set according to the IDH1-mutation status and 1p19q-codeletion status are summarized in On-line Table 2.
Interrater Agreement
Interrater analysis showed significant agreement in all VASARI imaging features. Interrater agreement for all the imaging features was good to excellent (κ value = 0.715–1.000) (On-line Table 3).
Significant Imaging Features in Differentiating IDH-Mutant and IDH-Wild Type LGGs (WHO Grade II and III Gliomas)
Various imaging features were significantly different between the IDH1-wild type and IDH1-mutant groups according to the Student t test and χ2 tests (On-line Table 4). The significantly different features included the major axis length, tumor location, side of the tumor epicenter, presence of enhancement, proportion of enhancing tumors, proportion of edema, proportion of necrosis, cysts, multifocal/multicentric distribution, infiltrative tumors, solid tumor enhancement, enhancing margin, nonenhancing margin, diffusion characteristics, pial invasion, ependymal extension, cortical involvement, and deep white matter invasion. Among them, 4 factors were independently associated with predicting the IDH1 mutation by the LASSO procedure (On-line Fig 2), including the nonlobar tumor location, proportion of enhancing tumors of >33%, multifocal/multicentric distribution, and definition of the nonenhancing margin (Table 1). The IDH1-mutant group had a lobar tumor location and smaller proportion of enhancing tumors (Fig 1). However, the IDH1-wild type group had multifocal/multicentric distribution and poor definition of the nonenhancing margin. The AUC for the optimal model was 0.859 (95% confidence interval, 0.784–0.934). When the 4 parameters were used for diagnosis in the independent validation set, it reached an AUC of 0.778 (95% confidence interval, 0.619–0.893).
Prediction for an IDH1-mutation status in lower grade gliomas using the LASSO procedure
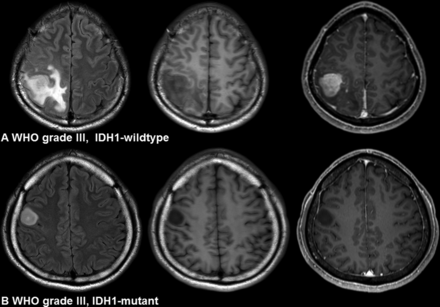
Imaging characteristics of IDH1-wild type and IDH1-mutant gliomas. A, A 24-year-old woman with an IDH1-wild type lower grade glioma (anaplastic astrocytoma, World Health Organization grade III). The imaging features are as follows: a lobar location, proportion of enhancing tumor of >33%, and poorly defined nonenhancing margin. B, A 42-year-old man with an IDH1-mutant lower grade glioma (anaplastic astrocytoma, WHO grade III). The imaging features are as follows: a lobar location with focal distribution, proportion of enhancing tumor of <33%, and well-defined nonenhancing margin.
Significant Imaging Features in Differentiating IDH-Mutant and IDH-Wild Type Gliomas in WHO Grade II Gliomas
Various imaging features were significantly different between the IDH1-wild type and IDH1-mutant WHO grade II subgroups according to the Student t test and χ2 test results (On-line Table 5). Among them, 4 factors were independently associated with predicting the IDH1 mutation by the LASSO procedure. Significant differences were noted between the IDH1-wild type group and IDH1-mutant group, including the side of the tumor epicenter (central), multifocal/multicentric distribution, pial invasion, and ependymal involvement (Table 1). The AUC for the optimal model was 0.830 (95% CI, 0.753–0.907).
Significant Imaging Features in Differentiating IDH-Mutant and IDH-Wild Type Gliomas in WHO Grade III Gliomas
Various imaging features were significantly different between the IDH1-wild type and IDH1-mutant WHO grade III subgroups according to the Student t test and χ2 test results (On-line Table 6). Among them, 4 factors were independently associated with predicting the IDH1 mutation by the LASSO procedure. Significant differences were noted between the IDH1-wild type group and IDH1-mutant group, including the nonlobar tumor location, proportion of enhancing tumors of >33%, multifocal/multicentric diffusion, and cortical involvement (Table 1). The AUC for the optimal model was 0.873 (95% CI, 0.794–0.952).
Significant Imaging Features in Differentiating the IDH-Mutant, No 1p/19q-Codeletion and IDH-Mutant, 1p/19q-Codeleted Groups
The frequency of pial invasion and diffusion characteristics was significantly different according to the 1p/19q-codeletion status in the IDH1-mutant subgroup (P = .039 and .020, respectively) (On-line Table 7). The IDH1-mutant, 1p/19q-codeleted group had significantly more pial invasion and a mixed pattern of high and intermediate ADC values or restricted diffusion characteristics than the IDH-mutant, no 1p/19q-codeletion group (Fig 2).
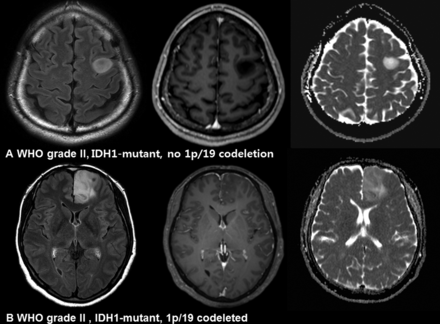
Imaging characteristics of IDH1-mutant, no 1p/19q-codeletion and IDH1-mutant, 1p/19q-codeleted gliomas. A, A 34-year-old man with an IDH1-mutant, no 1p/19q-codeletion glioma (diffuse astrocytoma, World Health Organization grade II). The nonenhancing T2 hyperintense mass shows increased diffusivity. B, A 49-year-old woman with an IDH1-mutant, 1p/19q-codeleted glioma (oligodendroglioma, WHO grade II). The infiltrative T2 hyperintense mass shows a mixed pattern of high and intermediate ADC values.
Discussion
In our study, we comprehensively analyzed the MR imaging features of VASARI according to the molecular subtypes of LGGs. A noninvasive prediction of IDH mutation is important; a recent study suggested that in IDH1-mutant gliomas, maximal surgical resection, including enhancing and nonenhancing tumors, may contribute to a better prognosis.12 In contrast, a survival benefit was noted in the complete resection of only enhancing tumors of IDH1-wild type gliomas, whereas no survival benefit was observed in further resection of the nonenhancing tumor portion. Although maximal resection of total tumor volume remains the optimal treatment, inference can be made about the contrast-enhancing portion of tumors and accessibility based on IDH1-mutation status in planning and performing an operation.13 The overall survival, progression-free survival, and response to chemoradiotherapy are different according to the IDH-mutation status3,14,15; therefore, preoperative prediction of the molecular classification of LGGs is useful to guide the treatment decision and predict the prognosis. In addition, a selective inhibitor impaired the growth of IDH1-mutant glioma cells,16 and the noninvasive prediction of IDH1 mutation could assist in the development of treatment strategies such as targeted therapy.
Our proposed approach can be applied by visual assessment of conventional MR imaging, which is practical for implementation and economical. Overall, IDH1-mutant gliomas exhibited less invasive imaging features compared with IDH-wild type gliomas. All IDH1-mutant gliomas presented with a unilateral epicenter and focal distribution. With the 4 significant imaging features selected by the LASSO procedure, which are the nonlobar location, proportion of the enhancing tumor, multifocal/multicentric distribution, and definition of the nonenhancing margin, AUCs were 0.859 in the discovery set and 0.778 in the validation set in our study. In the WHO grade II and III subgroups, the significant imaging features according to the IDH1-mutation status mostly overlapped, except that pial invasion and ependymal extension were also significant factors in WHO grade II gliomas, and the tumor location, proportion of enhancement, and definition of the enhancing margin were significant factors in WHO grade III gliomas. A recent study showed that the proportion of necrosis and lesion size predicted the IDH1-mutation status, which correlates with our results.17 In WHO grade III gliomas, IDH-mutant tumors are strongly associated with a frontal location,18⇓–20 whereas IDH-wild type tumors are frequently located outside the frontal lobe.19,20 The predominant frontal lobe location of IDH-mutant gliomas may be because neuroglial progenitor cells in the forebrain subventricular zone are likely cells of origin for IDH-mutant gliomas.21,22 In another study with WHO grade II gliomas, IDH-wild type tumors had greater tumor volume and an infiltrative pattern on MR images.23 Moreover, in WHO grade II and III astrocytomas, IDH-mutant gliomas were predominantly located in a single lobe and had less contrast enhancement,24 which are in accordance with our results. In glioblastomas, IDH1-wild type tumors showed a higher proportion of enhancing tumor,25 and IDH1-mutant tumor had a less invasive phenotype and frontal lobe predominance,25⇓–27 which is like our findings in LGGs.
Conflicting results have been reported regarding the association between the IDH mutation and tumor borders.24,28 These discordant results may be because in 1 study, the margins of the nonenhancing and enhancing portions were not differentiated,24 whereas in our study, IDH1-mutant tumors frequently showed a poorly defined enhancing margin and a well-defined nonenhancing margin, which correlated with the results of another study.28 In many patients with IDH1-mutant tumors, there was no enhancement, and if enhancement was present, it was faint, thereby displaying a poorly defined enhancing margin. In terms of the nonenhancing margin, a recent study also showed that grade II and III astrocytomas with IDH mutation had sharp borders on T2-weighted imaging,29 which is consistent with our results. In WHO grade II and III astrocytomas, IDH1-wild type tumors showed lower apparent diffusion coefficient values than IDH1-mutant tumors, concordant with our results.28,30
Increased cell proliferation or cellularity decreases ADC values in glioblastomas.13,31 This feature may suggest that IDH mutations can decrease glioma proliferation, and it explains why an IDH mutation is a favorable prognostic marker in patients with gliomas.2,32 Pial invasion and ependymal extension were also significant factors in WHO grade II gliomas in our study, according to the IDH1-mutation status. In the previous literature, there was no discussion of the imaging findings of pial invasion, possibly because these findings were not included in the analyses. The increased incidence of pial invasion in IDH-wild type gliomas may be due to the invasiveness of IDH-wild type gliomas, but further study is indicated to validate the finding. Recent studies have shown that glioblastomas with ependymal extension showed a significant decrease in overall survival.33 This may be because tumors arising from neural stem cells in the subventricular zone have a higher potential for invasiveness, which may be correlated with ependymal involvement.34
LGGs with an IDH mutation and 1p/19q codeletion are associated with favorable outcomes and have sensitivity to chemotherapy with alkylating agents.2,3,35⇓–37 Several studies have analyzed the association between a 1p/19q codeletion and MR imaging features.17,19,20,28,38⇓–40 In our study, IDH1-mutant, 1p/19q-codeleted gliomas more frequently had a mixed pattern of high and intermediate ADC values or restricted diffusion characteristics and more pial invasion compared with IDH1-mutant, no codeletion gliomas. There has been controversy regarding the association between a 1p/19q codeletion and tumor borders.19,20,28,39 Other studies have reported a low accuracy for predicting 1p/19q codeletion using conventional and advanced MR imaging.28,40 The frequent mixed pattern of high and intermediate ADC values or restricted diffusion characteristics in IDH1-mutant, 1p/19q-codeleted gliomas may reflect the difference in oligodendroglial tumor biology compared with other gliomas.41
Several studies have shown discordant results about the association between the ADC and 1p/19q status,28,40 which may be because the evaluation was performed in different study groups. Previous studies included patients with oligodendroglial tumors based on histopathology findings regardless of the IDH-mutation status; then, the authors classified them into groups with and without 1p/19q codeletion. Therefore, a small number of patients with IDH-wild type gliomas were included in those studies.20,28,39 However, our study analyzed the association between imaging features and the 1p/19q codeletion in an IDH1-mutant subgroup, and the different inclusion criteria may have partially contributed to discordant results. Further study is needed to validate our results in a larger population with a homogeneous group.
For reproducible and reliable evaluation of imaging features, imaging analysis was performed on the basis of VASARI MR imaging features.6 A controlled set of imaging features (ie, VASARI) has many benefits, including good-to-excellent interrater agreement. A controlled lexicon will facilitate more concrete knowledge regarding the relationship of imaging features with clinical and genotypic features and improve the communication of results among clinicians.7 Several studies have used the VASARI lexicon to evaluate imaging features, and 1 study found that invasive imaging characteristics assessed with VASARI were associated with upregulation of the oncogene in glioblastomas.25,26
Our study has several significant limitations. First, it was based on a single-center, retrospectively collected dataset. Second, prognostic markers were not analyzed because patients with LGGs have relatively long overall survival, especially with WHO grade II gliomas. Further studies are necessary to correlate prognostic markers such as overall survival and progression-free survival with genotypic and imaging features. Third, there is the possibility of biopsy sampling error in cases of stereotactic biopsy or subtotal/partial resections, as previously reported.31 Fourth, several imaging features noted as significant in the LASSO procedure in predicting IDH-wild type in WHO grade II gliomas showed a relatively low incidence, as seen in On-line Table 5; therefore, features may have limited value for clinical application. Considering the low incidence of IDH mutation in grade II gliomas, further studies with larger populations are necessary to find noninvasive imaging biomarkers with detailed imaging feature analysis in WHO grade II gliomas.
Conclusions
Preoperative MR imaging phenotypes are different according to the molecular markers in LGGs, and they may be helpful in predicting the IDH1-mutation status. The imaging phenotypes of nonlobar location, proportion of enhancing tumor, multifocal/multicentric distribution, and poor definition of nonenhancing margin assist in predicting IDH1-wild type LGGs.
Footnotes
Disclosures: Yae Won Park—RELATED: Grant: Yonsei University College of Medicine (6-2015-0079), Comments: faculty research grants of the Yonsei University College of Medicine (6-2015-0079)*. *Money paid to the institution.
S.S. Ahn was supported by faculty research grants of the Yonsei University College of Medicine (6-2015-0079).
References
- Received May 26, 2017.
- Accepted after revision August 14, 2017.
- © 2018 by American Journal of Neuroradiology