Abstract
BACKGROUND AND PURPOSE: Precision medicine is an approach to disease diagnosis, treatment, and prevention that relies on quantitative biomarkers that minimize the variability of individual patient measurements. The aim of this study was to assess the intersite variability after harmonization of a high-angular-resolution 3T diffusion tensor imaging protocol across 13 scanners at the 11 academic medical centers participating in the Transforming Research and Clinical Knowledge in Traumatic Brain Injury multisite study.
MATERIALS AND METHODS: Diffusion MR imaging was acquired from a novel isotropic diffusion phantom developed at the National Institute of Standards and Technology and from the brain of a traveling volunteer on thirteen 3T MR imaging scanners representing 3 major vendors (GE Healthcare, Philips Healthcare, and Siemens). Means of the DTI parameters and their coefficients of variation across scanners were calculated for each DTI metric and white matter tract.
RESULTS: For the National Institute of Standards and Technology diffusion phantom, the coefficients of variation of the apparent diffusion coefficient across the 13 scanners was <3.8% for a range of diffusivities from 0.4 to 1.1 × 10−6 mm2/s. For the volunteer, the coefficients of variations across scanners of the 4 primary DTI metrics, each averaged over the entire white matter skeleton, were all <5%. In individual white matter tracts, large central pathways showed good reproducibility with the coefficients of variation consistently below 5%. However, smaller tracts showed more variability, with the coefficients of variation of some DTI metrics reaching 10%.
CONCLUSIONS: The results suggest the feasibility of standardizing DTI across 3T scanners from different MR imaging vendors in a large-scale neuroimaging research study.
ABBREVIATIONS:
- AD
- axial diffusivity (units of 10−3 mm2/s)
- CoV
- coefficient of variation
- FA
- fractional anisotropy
- MD
- mean diffusivity (units of 10−3 mm2/s)
- NIST
- National Institute of Standards and Technology
- PVP
- polyvinylpyrrolidone
- RD
- radial diffusivity (units of 10−3 mm2/s)
- TBI
- traumatic brain injury
- TRACK-TBI
- Transforming Research and Clinical Knowledge in Traumatic Brain Injury
The new paradigm of precision medicine relies on quantitative biomarkers validated for their accuracy and efficacy. They must also be standardized so that precise and reproducible measurements can be made despite differences in instrumentation and procedures.1 Medical imaging can provide many clinically useful biomarkers; however, the precision of its metrics must first be established in large-scale multisite studies that encompass the range of scanning hardware and software used to acquire the data.
DTI studies of patients with traumatic brain injury (TBI) show alterations of white matter microstructure in many tracts, across the range of severe, moderate, and mild TBI.2⇓⇓–5 However, TBI is a highly heterogeneous pathology in cause, severity, and clinical course. New standardized clinical and imaging biomarkers are needed to enhance outcome prediction and for triage to the most appropriate therapeutic interventions. Transforming Research and Clinical Knowledge in TBI (TRACK-TBI), a National Institutes of Health–funded study, began in October 2013 with the goal of enrolling 3000 patients with TBI at 11 enrollment sites across the United States. The objective was to create a large, high-quality data base that integrates clinical, imaging, proteomic, genomic, and outcome measures to establish more precise methods for TBI diagnosis and prognosis.6 The first critical element for a multicenter imaging study is to minimize the intersite variability. Biomarkers derived from DTI metrics must be optimally robust with respect to differences in the acquisition hardware and software across medical centers.7 Intersite differences can originate from variations in scanner manufacturer; hardware characteristics such as field strength, gradient strength, and speed; and the type of radiofrequency coil, software version, site-specific quality control procedures, and adherence to the research protocol.8
Diffusion phantom and human volunteer studies are needed to assess the reproducibility and variability of the imaging data. Previous studies have shown DTI variability on test-retest studies,9⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓–20 but only 3 previous articles7,21,22 have studied systematic DTI variability due to intrinsic differences between different MR imaging systems in an isotropic phantom and in a human volunteer, either within a single site or across sites. However, these prior investigations were limited to relatively few sites in the case of multicenter studies, to 1.5T scanners or a combination of 1.5T and 3T scanners, to a DTI protocol with low angular resolution, and, in 1 case, to nonharmonized DTI by using different protocols at different sites.
In this study, we assessed the intersite precision of DTI metrics from a harmonized high-angular-resolution DTI protocol across thirteen 3T scanners at the 11 sites enrolling patients for the TRACK-TBI study. High-angular-resolution diffusion imaging, which requires the acquisition of ≥30 diffusion gradient directions per scan, has been proposed to improve the accuracy and reliability of DTI metrics compared with standard DTI acquisitions with fewer directions.23,24 In addition to the traveling human volunteer, data were acquired from a novel isotropic diffusion phantom developed at the National Institute of Standards and Technology (NIST), which allows precise assessment of the range of diffusivities that might be encountered in healthy and pathologic brain tissue.
Materials and Methods
All study procedures were approved by the institutional review boards of all 11 enrollment sites of the TRACK-TBI multicenter study.
The initial diffusion phantom and human brain data were acquired from all 13 scanners at the 11 sites within a 4-month time period at the beginning of the study. The acquisition, processing, and analysis of the isotropic diffusion phantom data were performed by a PhD MR imaging physicist (A.J.M), who is a Professor of Radiology with 25 years of experience in industry and academia. The traveling volunteer brain imaging data were processed and analyzed by a PhD scientist (E.M.P.) with 8 years of experience in diffusion tensor image processing and analysis in TBI. All of the imaging analysis was overseen by a board-certified neuroradiologist (P.M.) with 20 years of experience with DTI research and who is the director of the imaging core and a Principal Investigator of the multicenter study.
NIST Isotropic Diffusion Phantom
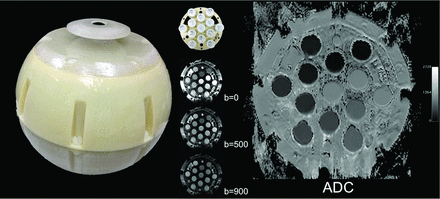
Diffusion MR imaging was acquired from a prototype isotropic diffusion phantom in a 3D-printed shell developed at the NIST and from the brain of a traveling healthy volunteer (male, 49 years of age) on thirteen 3T MR imaging scanners at 11 different sites by using 8- or 12-channel head radiofrequency coils. Isotropic phantoms are used to calibrate diffusivities, so a full DTI acquisition or identical parameters with the traveling volunteer sequences is not a requirement. The main objective of the isotropic phantoms is to verify the precision of the principal axis diffusivities because all DTI metrics are computed from these diffusivities.
The diffusion phantom was scanned at b=0, 500, and 900 s/mm2 with 1.1-mm in-plane resolution and 5-mm sections, by using 3 orthogonal directions and 4 signal averages. An 11-section protocol was used and the image stack was prescribed such that the sections were centered on the phantom and intersected vials within the phantom cross-sectionally.
Traveling Volunteer
The traveling volunteer was scanned once at each center during the same session as the NIST phantom. All volunteer scans were acquired within a 4-month interval. Additionally, from one of the sites (site 3), 2 DTI scans were acquired on the same day from the human traveling volunteer, 30 months after the first DTI acquisition from this volunteer. The characteristics of the scanners for each site and parameters of acquisition from the traveling volunteer in each site are reported in Table 1. The DTI protocol is based on the “Enhanced DTI” standard established by the Alzheimer Disease Neuroimaging Initiative 2 study for GE Healthcare scanners (http://adni.loni.usc.edu/wpcontent/uploads/2010/05/ADNI2_GE_22_E_DTI.pdf), except that 64 diffusion directions were used at most sites instead of 60 directions to accommodate the Siemens in-product DTI sequence in specifying the number of directions. In brief, the protocol consists of a multislection spin-echo echo-planar sequence with 2.7-mm isotropic spatial resolution and a b-value of 1300 s/mm2, with 8 additional brain volumes acquired at b=0 s/mm2.
DTI acquisition protocol for the 11 academic medical centersa
All the images were inspected, and none had any major artifacts. One of the advantages of using high-angular-resolution diffusion imaging is that a corrupted image in any single direction has a minimal effect on the fitting or metrics. Thus, images from high-angular-resolution diffusion imaging produce more accurate and reliable results than lower angular resolution acquisitions.
Preparation and Processing of the NIST Isotropic Diffusion Phantom
We established an array of thirteen 20-mL vials containing variable amounts of the polymer polyvinylpyrrolidone (PVP) (Fig 1).25 PVP containing vials, with mass fractions of 0%, 10%, 20%, 30%, 40%, and 50% were arranged in a plane, with 0% PVP (deionized water) at the center and increasing concentrations of PVP in 2 rings around the central vials. Vials were contained in a spheric water-tight vessel. The phantom was transported between sites in a dry state in a foam enclosure. On arrival at each site, the phantom was initially packed with ice and placed in a refrigerator. In parallel, a slurry of ice and water was prepared and allowed to equilibrate for 1–2 hours. Ice water was then added to the phantom along with as much additional ice as could be accommodated; the phantom was returned to the refrigerator and left overnight (8–10 hours). In the morning, additional ice was packed into the phantom and left in an insulated container until imaged (within 2 hours).
Prototype isotropic diffusion phantom in a 3D-printed shell from the National Institute of Standards and Technology.
DTI Preprocessing and Analysis for the Traveling Volunteer
DTI preprocessing and analysis were performed by using tools from the Oxford Centre for Functional MR Imaging of the Brain (FMRIB) Software Library (FSL (http://www.fmrib.ox.ac.uk/fsl)).
First, images were corrected for eddy current distortions and motion by using an average of the 8 b=0 s/mm2 volumes as a reference. The registered images were skull-stripped by using the FSL Brain Extraction Tool (http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/BET). All the resulting brain masks were visually inspected. Fractional anisotropy (FA) maps were calculated by using the FMRIB Diffusion Toolbox (http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FDT).26 After calculation of the FA map, a voxelwise statistical analysis of the FA data was performed by using Tract-Based Spatial Statistics (TBSS; http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/TBSS).27,28 Within the TBSS pipeline, FA data were aligned into the common FMRIB58 FA template, which is in Montreal Neurological Institute 152 atlas standard space, by using the nonlinear registration algorithm FMRIB Nonlinear Registration Tool (FNIRT; http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FNIRT).29
Next, a mean FA image was created from the images for all the subjects' serial scans at different sites in this common space and thinned to generate a mean FA white matter skeleton that represented the center of all tracts common to the entire group of scans. The FA white matter skeleton was thresholded to FA > 0.2 to exclude gray matter and voxels containing partial volume effects with gray matter. This process ensured that each dataset skeleton was in the group space while also representing the center of the subject's unique white matter bundles. The aligned FA volume was then projected onto the skeleton by filling the skeleton with FA values from the nearest relevant tract center. This was achieved for each skeleton voxel by searching perpendicular to the local skeleton structure for the maximum value in the FA image of the subject. Mean FA, mean diffusivity (MD), axial diffusivity (AD), and radial diffusivity (RD) values were obtained from the FA skeleton map of each scan.
Specific fasciculi were studied by using masks obtained from the FSL Jülich histologic atlas (http://neuro.debian.net/pkgs/fsl-juelich-histological-atlas.html) mapped on the standard Montreal Neurological Institute space and resampled to 1-mm resolution. Binary mask images from the fasciculi of interest were used to mask the individual FA (skeletonized) maps previously registered to the Montreal Neurological Institute standard space by using the nonlinear tools in the TBSS procedure. We included 15 large main fasciculi. Mean FA, MD, AD, and RD values were obtained from each subject's white matter skeleton and each of the skeletonized ROIs.
Statistical Analysis
NIST Isotropic Diffusion Phantom.
ADC images were reconstructed, and ROI analysis was performed. ADC values were quantified in the center section, which corresponded to the central plane of the phantom. A circular ROI measuring 1.2–1.3 cm2 was placed in the center of each of the 13 vials, and the mean ADC value was determined. A mean ADC and its coefficient of variation (CoV), which is obtained from calculating the SD divided by the average, were established for each vial across all sites.
Traveling Volunteer.
The CoV was calculated for each DTI metric in the whole white matter skeleton and for each fasciculus to summarize the amount of variation across scanners at the 11 sites. Furthermore, we also calculated a normalized CoV for each of the preselected ROIs, in which each DTI metric was divided by its corresponding mean value per site, as has been performed in prior DTI studies of TBI.30,31
Results
NIST Isotropic Diffusion Phantom
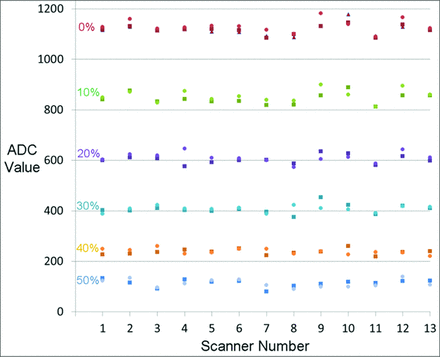
There were 3 ADC measures for the three 0% PVP vials and 2 measures each for the paired 10%, 20%, 30%, 40%, and 50% PVP vials. The differences between the inner and outer rings of vials were not statistically significant (P > .05). Figure 2 shows the consistency of ADC measurements within and across systems. Table 2 provides the mean, SD, and CoV for each concentration of PVP. Data are shown for the full set of TRACK-TBI systems and for a subset of systems that shared the same manufacturer and model (Siemens Trio). The latter is the most common scanner type in TRACK-TBI study, with no more than 2 of any other system. The CoV is <3.8% for concentrations of PVP of <30% w/w. The CoV is slightly decreased if a single scanner type is considered. At 40% and 50% w/w PVP, the diffusivity decreased and the resulting CoV became larger.
ADC measurements obtained across all 13 scanners in TRACK-TBI. The measured ADC in units of 10−6 mm2/s for vials containing 0%–50% PVP are plotted for each TRACK-TBI MR imaging system. The NIST phantom allows 3 measures of 0% PVP and 2 measures of each of the 10%–50% PVP concentrations. Data are shown for the central vial (triangle), inner vials (squares), and outer vials (circles) of the phantom.
Mean ADC, SD, and CoV for each concentration of PVP
Traveling Volunteer
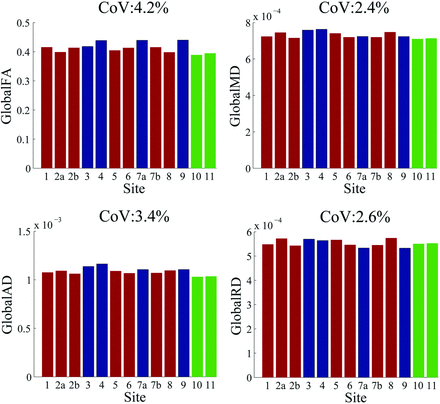
Whole-brain DTI measurements across sites are shown in Fig 3. The CoV for global white matter skeleton DTI parameters within and across scanners was <5%, showing consistency comparable with the NIST diffusion phantom results. Table 3 and Fig 4 report the mean of the DTI parameters of each of the 14 white matter tracts (left and right sides) and the CoV extracted from the DTI parameters and their normalized values. For FA, MD, AD, and RD values, the CoV was ≤5% for most tracts both within and across scanners (Tables 3 and 4). The CoV of normalized DTI values was less than that of non-normalized DTI values.
Global non-normalized DTI parameters and CoV across sites. Red indicates Siemens; Blue, GE Healthcare; Green, Phillips Healthcare. By order of sites, see Table 1.
Mean DTI parameters across all 13 scanners and their CoV
Graphic representation of CoVs across all 13 scanners. The coefficient of variation (percentage) is in orange. Mean non-normalized DTI parameters are in gray. r indicates right; l, left; SLF, superior longitudinal fasciculus; SFO, superior fronto-occipital fasciculus; CST, corticospinal tract; CC, corpus callosum; CING, cingulum corticospinal tract; ACR, anterior corona radiata; ALIC, anterior limb of internal capsule; PCR, posterior corona radiata; PLIC, posterior limb of internal capsule; PTR, posterior thalamic radiation; EC, external capsule; UNC, uncinate fasciculus.
Intrascanner DTI measures for site 3 and their CoV
Discussion
In this study of intersite variability of DTI by using both an isotropic phantom and a human volunteer, the 3T high-angular-resolution diffusion imaging protocol yielded CoVs below 5% for FA, MD, AD, and RD, averaged over the white matter skeleton of the whole brain. This finding demonstrates acceptably low intersite variation despite the 3 different scanner vendors and 6 different scanner models across the 11 centers. When examining the DTI parameters for specific tracts, we found better measurement precision in large central tracts than in small peripheral white matter, with reduced variability (lower CoV) when normalizing the values by the global DTI parameters per site. The low intersite variability of diffusion metrics was confirmed by ADC results across all 13 scanners from the NIST diffusion phantom over a range of values characteristic of the healthy human brain, as well as high and low values caused by pathology such as TBI.
The NIST has developed a series of MR imaging isotropic diffusion phantoms to enable stable measurements to mimic human tissue responses to MR imaging in a predictable and repeatable way for use in calibrating MR imaging scanners. The NIST isotropic diffusion phantom in the present study contained variable amounts of the PVP, which, at different concentrations, has shown properties similar to those of healthy brain tissue.25 The different concentrations in each of the vials corresponded with the expected degrees of diffusivity and showed minimal variation between scanners. The relatively poor CoV seen at very low diffusivities likely comes from the selected range of b-values, which were limited to values of 0, 500, and 900 s/mm2. Incorporation of a larger b-value is likely to substantially improve the variance in high-PVP-concentration vials, and we have recently adapted our protocol to include a b=2000 s/mm2 value as well.
The results of our study, with CoVs ranging from 2% to 5%, generally concur with those of prior investigations of intersite DTI measurements in healthy subjects.9,10,18⇓–20 In a very recent study, Kamagata et al17 scanned 7 volunteers at 2 sites, both with 3T Achieva scanners (Philips Healthcare). With several analytic methods, including voxelwise comparison with TBSS, atlas-based analysis, and tract-specific analysis, they reported a CoV intersite variability of <4% in the case of tract-specific analysis and a CoV of <6% by using the atlas-based analysis. Even if the parameters of acquisition, scanners, and approaches of all these studies vary, the CoV seems to be a reliable and stable measure for intersite variation. However, all these studies compared groups of different healthy subjects, adding confounding factors due to intersubject biologic variability. In our study, this large source of variation was eliminated by using the same subject across all 13 scanners.
MR imaging phantoms enable more precise measurement of diffusivity values against known standards than is possible by using a living human brain. To our knowledge, only 3 multisite studies have previously assessed the intersite reliability of DTI measures in both a traveling volunteer and an isotropic diffusion phantom. In the most recent study, Grech-Sollars et al22 assessed the reproducibility of DTI in an ice water phantom and in 9 healthy volunteers across 8 scanners (1.5T Avanto; 1.5T Symphony; Siemens; 3T Achieva) at 5 sites, all by using different DTI acquisition parameters. The phantom used in that study consisted of 5 tubes filled with distilled water and 1 tube with sucrose. Despite the considerable variability across scanners and DTI protocols, the authors reported robustness across 1.5T and 3T scanners with CoVs computed from the non-normalized DTI values ranging from 1% to 7.4%.
Zhu et al21 evaluated the intra- and intersite differences in DTI measurements in 3 centers, all with 1.5T GE Healthcare Signa scanners, in a travelling human volunteer and a phantom fabricated with cells in a cylindric polycarbonate container filled with 3 chemicals (cyclic alcanes). Their normalized bias scores results suggested that the intersite variation, though relatively small among scanners of the same vendor, significantly affects DTI measurement accuracy and precision. In Walker et al,8 the investigators proposed a 2-step analysis for assessing phantom data in multisite DTI studies. These results were part of a pediatric brain development study at 5 sites with 1.5T scanners and the American College of Radiology MR imaging–accreditation phantom, consisting of 10 mmol of nickel chloride and sodium chloride. The results suggested that initial outlier identification is important for accurate assessment of inter- and intrasite variability and for identification of problems with data acquisition.
Our study offers several advances over these previous multisite DTI standardization studies. The novel NIST diffusion phantom has a much larger and more granular range of diffusivities, from 0.1 to 1.1 mm2/s with 6 distinct ADC values, compared with phantoms used in prior reports. This feature allows more precise calibration of the DTI values expected in healthy and pathologic brain tissue. We restricted the study to 3T scanners to avoid systematic differences due to field strength and because the much higher signal-to-noise ratio compared with 1.5T scanners enables measurement of DTI metrics such as FA with much less uncertainty.18 Our use of a harmonized high-angular-resolution diffusion imaging protocol with 60+ diffusion directions also allows more precise quantitation of DTI metrics.23,24 A prior multicenter DTI standardization study of 2 traveling volunteers in five 3T scanners (3 Trio scanners and 2 Signa scanners) also used a high-angular-resolution diffusion imaging protocol with 33 diffusion directions and found good concordance of FA and MD values across sites.19 Consistent with our results, these authors concluded that the comparability of DTI measures between different magnets supports the feasibility of multisite clinical trials by using DTI as an outcome measure. However, they reported greater intersite variability in DTI metrics than we found, with an average white matter CoV of 6.8% for FA and 4.1% for MD.
Despite having many more scanners (13 versus 5) and many more scanner models (6 versus 2) than Fox et al,19 we found a global white matter CoV of 4.2% for FA and 2.4% for MD. This finding is likely due to the hand-drawn ROI measurements of Fox et al, which introduce intrarater and interrater variability, compared with the fully automated DTI measurements of the present study using TBSS, which has the added advantage of evaluating white matter throughout the entire brain instead of only a few selected regions. In addition, when estimating the FA CoV of a specific tract (ie, the anterior corona radiata) from a prior DTI study of mild TBI,5 we found a 5% CoV for the anterior corona radiata, known to be commonly injured after mild TBI,2 across the pooled group of patients with complicated mild TBIs and controls, with a statistically significant mean difference of 3% between patient and control groups at P < .05. Thus, the interscanner variation of FA observed in this tract in the current study across 13 different 3T scanners of various types is similar to the intersubject variability of FA on a single scanner.5
A limitation of our study is that only a single volunteer could be assessed at all sites, given the travel expenses involved in flying multiple volunteers to be scanned at 11 different medical centers located throughout the United States. Also, intrasite scan-rescan reliability was obtained at only 1 site, though with the same human subject scanned across all of the sites. Our results indicate, as expected, that the variability of DTI metrics is less within the same scanner (Table 4) than across different scanners (Table 3). There were small variations in the DTI protocol performed at each site due to hardware or software idiosyncrasies of each vendor platform; however, low intersite variability was achieved despite these minor protocol differences. This study was designed to measure the magnitude of variability, but the many possible sources of variation due to scanner hardware and software factors, as well as dozens of pulse sequence parameters, are beyond the scope of this study. This subject remains an open area in the DTI literature, which requires future investigation. The application of newly developed methods to retrospectively compensate for intersite differences in diffusion MR imaging acquisition by using correction factors may further reduce the variability of DTI metrics in multisite studies and improve reproducibility,20 including for longitudinal data.32 This application is particularly important for the investigation of smaller peripheral white matter tracts in which intersite variation is higher than in large central white matter regions.
Conclusions
By using a novel isotropic diffusion phantom and a traveling volunteer, we have demonstrated the feasibility of standardizing DTI on 3T scanners from all 3 major MR imaging vendors across the 11 patient enrollment sites of the TRACK-TBI study. Our findings support the viability and reproducibility of large-scale multisite DTI projects to validate diffusion-based biomarkers for TBI and many other neurologic and psychiatric disorders in the coming era of precision medicine.
Acknowledgments
We gratefully acknowledge the MR imaging technologists and study coordinators from all sites of the TRACK-TBI study. We also acknowledge the pioneering efforts of the Quantitative Imaging Biomarkers Alliance of the Radiological Society of North America, especially their contributions to previous designs of an isotropic diffusion phantom that led to the development of the NIST diffusion phantom.
Data used in preparation of this article were obtained from the Transforming Research and Clinical Knowledge in Traumatic Brain Injury (TRACK-TBI) project. As such, there are investigators within TRACK-TBI that contributed to the design and implementation and/or provided data but did not participate in analysis or writing of this report. A complete listing of TRACK-TBI investigators can be found at: http://tracktbi.ucsf.edu/organization-and-leadership.
Footnotes
Disclosures: Alastair Martin—RELATED: Grant: National Institutes of Health, Comments: NIH U01NS086090 (Principal Investigator, Dr. Geoffrey T. Manley) “Transforming Research and Clinical Knowledge in Traumatic Brain Injury”*; Support for Travel to Meetings for the Study or Other Purposes: National Institutes of Health, Comments: NIH U01NS086090 (Principal Investigator, Dr. Geoffrey T. Manley) “Transforming Research and Clinical Knowledge in Traumatic Brain Injury.”* Michael A. Boss—UNRELATED: Patents (Planned, Pending, or Issued): National Institute of Standards and Technology, Comments: The phantom shell design was submitted for a patent, “MRI Phantom, Method for Making Same and Acquiring an MRI,” application No. 20150323639, The author has not received any payment related to this patent application.* Frank Ezekiel—RELATED: Grant: National Institutes of Health.* David M. Schnyer—RELATED: Grant: National Institute of Neurological Disorders and Stroke*; UNRELATED: Grants/Grants Pending: National Institute of Mental Health.* Christine L. MacDonald—RELATED: Grant: National Institutes of Health, Comments: 1U01NS086090-01 “Transforming Research and Clinical Knowledge in Traumatic Brain Injury”*; UNRELATED: Consultancy: Sinapis Pharma, Comments: I received payment for consultancy to Sinapis Pharma unrelated to this article; Employment: University of Washington, Comments: I am employed at the University of Washington; Grants/Grants Pending: R01HD083126, Comments: I received an RO1 grant from National Institutes of Health/National Institute of Neurological Disorders and Stroke for research unrelated to this article; Payment for Lectures including Service on Speakers Bureaus: University of Tennessee Health Sciences, Neurology Grand Grounds University of Hawaii, Pathology Chronic Traumatic Encephalopathy (CTE) Conference, Comments: I was an invited speaker to university and symposium lectures; I received payment for invited speaking engagements during the prior 36-month period unrelated to this article. Karen L. Crawford—RELATED: Grant: National Institutes of Health*; UNRELATED: Employment: University of Southern California; Grants/Grants Pending: National Institutes of Health, Alzheimer's Association, Michael J. Fox Foundation.* Arthur W. Toga—RELATED: Grant: National Institutes of Health*; UNRELATED: Employment: University of Southern California; Grants/Grants Pending: National Institutes of Health, Alzheimer's Association, Michael J. Fox Foundation.* Pratik Mukherjee—UNRELATED: Board Membership: GE-National Football League Medical Advisory Board, Comments: No payment has been received by me or my institution in the prior 36 months for my service on this board; Payment for Lectures including Service on Speakers Bureaus: GE Healthcare, Comments: Webinar. *Money paid to the institution.
This research was funded by National Institutes of Health grant U01 NS086090.
Grant funding support for investigators: A.J.M., NIH R01EB012031, NIH U01NS086090, NIH R21CA186140, NIH R01CA194533, Voyager Therapeutics, Quest Diagnostics Incorporated; E.L.Y., NIH U01 NS086090, DoD W81XWH-14–2-0176, D.M.S., DoD-Navy W911QY-14-C-0070, NIH U01NS086090, NIH R01AG046460; C.L.M., NIH R01HD083126, DoD PT108802-SC104833-S1, NIH U01NS086090, DoD W81XWH-14–2-0176; A.I., NIH R44 NS 081792, NIH U01NS086090; A.W.T., NIH R01MH094343, NIH P41EB015922, NIH U01NS086090; and P.M., NIH R24MH106096, NIH R01HD072074, NIH U01 NS086090, DoD W81XWH-14–2-0176, DoD W81XWH-13–1-0494, GE 114-2014-GES-0001.
The contribution of the NIST is not subject to copyright in the United States.
Indicates open access to non-subscribers at www.ajnr.org
References
- 1.
- 2.
- 3.
- 4.
- 5.
- 6.
- 7.
- 8.
- 9.
- 10.
- 11.
- 12.
- 13.
- 14.
- 15.
- 16.
- 17.
- 18.
- 19.
- 20.
- 21.
- 22.
- 23.
- 24.
- 25.
- 26.
- 27.
- 28.
- 29.
- 30.
- 31.
- 32.
- Received May 1, 2016.
- Accepted after revision October 10, 2016.
- © 2017 by American Journal of Neuroradiology